Cooperative assembly of p97 complexes involved in replication termination
- PMID: 36329031
- PMCID: PMC9633789
- DOI: 10.1038/s41467-022-34210-y
Cooperative assembly of p97 complexes involved in replication termination
Abstract
The p97 ATPase extracts polyubiquitylated proteins from diverse cellular structures in preparation for destruction by the proteasome. p97 functions with Ufd1-Npl4 and a variety of UBA-UBX co-factors, but how p97 complexes assemble on ubiquitylated substrates is unclear. To address this, we investigated how p97 disassembles the CMG helicase after it is ubiquitylated during replication termination. We show that p97Ufd1-Npl4 recruitment to CMG requires the UBA-UBX protein Ubxn7, and conversely, stable Ubxn7 binding to CMG requires p97Ufd1-Npl4. This cooperative assembly involves interactions between Ubxn7, p97, Ufd1-Npl4, and ubiquitin. Another p97 co-factor, Faf1, partially compensates for the loss of Ubxn7. Surprisingly, p97Ufd1-Npl4-Ubxn7 and p97Ufd1-Npl4-Faf1 also assemble cooperatively on unanchored ubiquitin chains. We propose that cooperative and substrate-independent recognition of ubiquitin chains allows p97 to recognize an unlimited number of polyubiquitylated proteins while avoiding the formation of partial, inactive complexes.
© 2022. The Author(s).
Conflict of interest statement
J.C.W. is a co-founder of MoMa therapeutics, in which he has a financial interest. The remaining authors declare no competing interests.
Figures



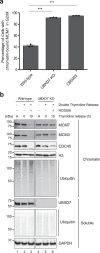

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

