Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays
- PMID: 36329085
- PMCID: PMC9633773
- DOI: 10.1038/s41598-022-23264-z
Discovering cell-active BCL6 inhibitors: effectively combining biochemical HTS with multiple biophysical techniques, X-ray crystallography and cell-based assays
Abstract
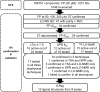
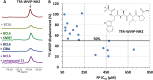
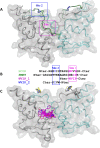
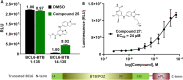
By suppressing gene transcription through the recruitment of corepressor proteins, B-cell lymphoma 6 (BCL6) protein controls a transcriptional network required for the formation and maintenance of B-cell germinal centres. As BCL6 deregulation is implicated in the development of Diffuse Large B-Cell Lymphoma, we sought to discover novel small molecule inhibitors that disrupt the BCL6-corepressor protein-protein interaction (PPI). Here we report our hit finding and compound optimisation strategies, which provide insight into the multi-faceted orthogonal approaches that are needed to tackle this challenging PPI with small molecule inhibitors. Using a 1536-well plate fluorescence polarisation high throughput screen we identified multiple hit series, which were followed up by hit confirmation using a thermal shift assay, surface plasmon resonance and ligand-observed NMR. We determined X-ray structures of BCL6 bound to compounds from nine different series, enabling a structure-based drug design approach to improve their weak biochemical potency. We developed a time-resolved fluorescence energy transfer biochemical assay and a nano bioluminescence resonance energy transfer cellular assay to monitor cellular activity during compound optimisation. This workflow led to the discovery of novel inhibitors with respective biochemical and cellular potencies (IC50s) in the sub-micromolar and low micromolar range.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

