ATP-binding and hydrolysis of human NLRP3
- PMID: 36329210
- PMCID: PMC9633759
- DOI: 10.1038/s42003-022-04120-2
ATP-binding and hydrolysis of human NLRP3
Abstract
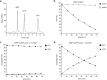
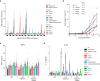
The innate immune system uses inflammasomal proteins to recognize danger signals and fight invading pathogens. NLRP3, a multidomain protein belonging to the family of STAND ATPases, is characterized by its central nucleotide-binding NACHT domain. The incorporation of ATP is thought to correlate with large conformational changes in NLRP3, leading to an active state of the sensory protein. Here we analyze the intrinsic ATP hydrolysis activity of recombinant NLRP3 by reverse phase HPLC. Wild-type NLRP3 appears in two different conformational states that exhibit an approximately fourteen-fold different hydrolysis activity in accordance with an inactive, autoinhibited state and an open, active state. The impact of canonical residues in the nucleotide binding site as the Walker A and B motifs and sensor 1 and 2 is analyzed by site directed mutagenesis. Cellular experiments show that reduced NLRP3 hydrolysis activity correlates with higher ASC specking after inflammation stimulation. Addition of the kinase NEK7 does not change the hydrolysis activity of NLRP3. Our data provide a comprehensive view on the function of conserved residues in the nucleotide-binding site of NLRP3 and the correlation of ATP hydrolysis with inflammasome activity.
© 2022. The Author(s).
Conflict of interest statement
The authors declare the following competing interests: M.G. and E.L. are co-founders and consultants of IFM Therapeutics. The other authors declare no competing interests.
Figures




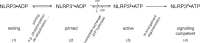
References
-
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. - PubMed
-
- Beutler B, et al. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu Rev. Immunol. 2006;24:353–389. - PubMed
-
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

