Complete mitochondrial genomes and updated divergence time of the two freshwater clupeids endemic to Lake Tanganyika (Africa) suggest intralacustrine speciation
- PMID: 36329403
- PMCID: PMC9635120
- DOI: 10.1186/s12862-022-02085-8
Complete mitochondrial genomes and updated divergence time of the two freshwater clupeids endemic to Lake Tanganyika (Africa) suggest intralacustrine speciation
Abstract
Background: The hydrogeological history of Lake Tanganyika paints a complex image of several colonization and adaptive radiation events. The initial basin was formed around 9-12 million years ago (MYA) from the predecessor of the Malagarasi-Congo River and only 5-6 MYA, its sub-basins fused to produce the clear, deep waters of today. Next to the well-known radiations of cichlid fishes, the lake also harbours a modest clade of only two clupeid species, Stolothrissa tanganicae and Limnothrissa miodon. They are members of Pellonulini, a tribe of clupeid fishes that mostly occur in freshwater and that colonized West and Central-Africa during a period of high sea levels during the Cenozoic. There is no consensus on the phylogenetic relationships between members of Pellonulini and the timing of the colonization of Lake Tanganyika by clupeids.
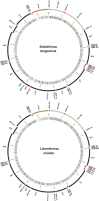
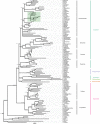
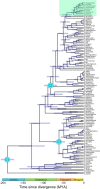
Results: We use short-read next generation sequencing of 10X Chromium libraries to sequence and assemble the full mitochondrial genomes of S. tanganicae and L. miodon. We then use Maximum likelihood and Bayesian inference to place them into the phylogeny of Pellonulini and other clupeiforms, taking advantage of all available full mitochondrial clupeiform genomes. We identify Potamothrissa obtusirostris as the closest living relative of the Tanganyika sardines and confirm paraphyly for Microthrissa. We estimate the divergence of the Tanganyika sardines around 3.64 MYA [95% CI: 0.99, 6.29], and from P. obtusirostris around 10.92 MYA [95% CI: 6.37-15.48].
Conclusions: These estimates imply that the ancestor of the Tanganyika sardines diverged from a riverine ancestor and entered the proto-lake Tanganyika around the time of its formation from the Malagarasi-Congo River, and diverged into the two extant species at the onset of deep clearwater conditions. Our results prompt a more thorough examination of the relationships within Pellonulini, and the new mitochondrial genomes provide an important resource for the future study of this tribe, e.g. as a reference for species identification, genetic diversity, and macroevolutionary studies.
Keywords: Clupeiformes; Great Lakes; Mitogenome; Phylogenetics; Time calibration.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Salzburger W, Van Bocxlaer B, Cohen AS. Ecology and evolution of the African Great Lakes and their faunas. Annu Rev Ecol Evol Syst. 2014;45(1):519–545.
-
- Cohen AS, Soreghan MJ, Scholz CA. Estimating the age of formation of lakes: an example from Lake Tanganyika, East African Rift system. Geology. 1993;21(6):511–514.
-
- Tiercelin J, Mondeguer A. The geology of the Tanganyika Trough. In: Coulter GW, editor. Lake Tanganyika and its life. Oxford: Oxford University Press; 1991. pp. 7–48.
-
- Tiercelin J-J, Lezzar K-E. A 300 million years history of rift lakes in central and east Africa: an updated broad review. In: Odada EO, Olago DO, editors. The East African great lakes: limnology, palaeolimnology and biodiversity advances in global change research. Springer: Dordrecht; 2002. pp. 3–60.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
