Early myeloid-derived suppressor cells accelerate epithelial-mesenchymal transition by downregulating ARID1A in luminal A breast cancer
- PMID: 36329699
- PMCID: PMC9623091
- DOI: 10.3389/fbioe.2022.973731
Early myeloid-derived suppressor cells accelerate epithelial-mesenchymal transition by downregulating ARID1A in luminal A breast cancer
Abstract
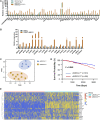
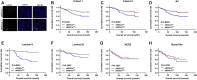
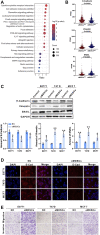
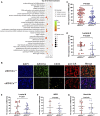
Early myeloid-derived suppressor cells (eMDSCs) are a newly characterized subclass of MDSCs, which exhibit more potent immunosuppressive capacity than classical MDSCs. Previously, we found high eMDSCs infiltration was correlated with poor prognosis of breast cancer, though the regulatory mechanisms have not been fully understood. Here, we constructed a 21-gene signature to evaluate the status of eMDSCs infiltration within breast cancer tissues and found that highly infiltrated eMDSCs affected the prognosis of breast cancer patients, especially in luminal A subtype. We also found that eMDSCs promoted epithelial-mesenchymal transition (EMT) and accelerated cell migration and invasion in vitro. Meanwhile, eMDSCs significantly downregulated ARID1A expression in luminal A breast cancer, which was closely associated with EMT and was an important prognostic factor in breast cancer patients. Moreover, significant changes of EMT-related genes were detected in luminal A breast cancer cells after co-cultured with eMDSCs or ARID1A knock-down and overexpression of ARID1A significantly reversed this procedure. These results implied that eMDSCs might suppress the ARID1A expression to promote EMT in luminal A breast cancer cells, which might provide a new light on developing novel treatment regimens for relapsed luminal A breast cancer after conventional therapies.
Keywords: ARID1A; breast cancer; early-stage myeloid-derived suppressor cells; epithelial-mesenchymal transition; prognostic factor.
Copyright © 2022 Chen, Li, Ji, Liu, Zhou, Xu, Wang, Li and Yu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources

