Multigene manipulation of photosynthetic carbon metabolism enhances the photosynthetic capacity and biomass yield of cucumber under low-CO2 environment
- PMID: 36330244
- PMCID: PMC9623318
- DOI: 10.3389/fpls.2022.1005261
Multigene manipulation of photosynthetic carbon metabolism enhances the photosynthetic capacity and biomass yield of cucumber under low-CO2 environment
Abstract
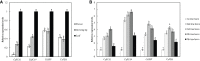
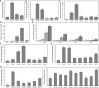
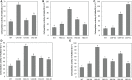
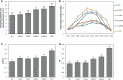
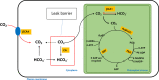
Solar greenhouses are important in the vegetable production and widely used for the counter-season production in the world. However, the CO2 consumed by crops for photosynthesis after sunrise is not supplemented and becomes chronically deficient due to the airtight structure of solar greenhouses. Vegetable crops cannot effectively utilize light resources under low-CO2 environment, and this incapability results in reduced photosynthetic efficiency and crop yield. We used cucumber as a model plant and generated several sets of transgenic cucumber plants overexpressing individual genes, including β-carbonic anhydrase 1 (CsβCA1), β-carbonic anhydrase 4 (CsβCA4), and sedoheptulose-1,7-bisphosphatase (CsSBP); fructose-1,6-bisphosphate aldolase (CsFBA), and CsβCA1 co-expressing plants; CsβCA4, CsSBP, and CsFBA co-expressing plants (14SF). The results showed that the overexpression of CsβCA1, CsβCA4, and 14SF exhibited higher photosynthetic and biomass yield in transgenic cucumber plants under low-CO2 environment. Further enhancements in photosynthesis and biomass yield were observed in 14SF transgenic plants under low-CO2 environment. The net photosynthesis biomass yield and photosynthetic rate increased by 49% and 79% compared with those of the WT. However, the transgenic cucumbers of overexpressing CsFBA and CsSBP showed insignificant differences in photosynthesis and biomass yield compared with the WT under low-CO2.environment. Photosynthesis, fluorescence parameters, and enzymatic measurements indicated that CsβCA1, CsβCA4, CsSBP, and CsFBA had cumulative effects in photosynthetic carbon assimilation under low-CO2 environment. Co-expression of this four genes (CsβCA1, CsβCA4, CsSBP, and CsFBA) can increase the carboxylation activity of RuBisCO and promote the regeneration of RuBP. As a result, the 14SF transgenic plants showed a higher net photosynthetic rate and biomass yield even under low-CO2environment.These findings demonstrate the possibility of cultivating crops with high photosynthetic efficiency by manipulating genes involved in the photosynthetic carbon assimilation metabolic pathway.
Keywords: carbon metabolism; cucumber; greenhouse; highphotosynthetic efficiency; low-CO2; multigene manipulation.
Copyright © 2022 Chen, Wang, Feng, Guo, Guan, Zhang, Li, Xing, Sun and Tan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Chen Z. F., Kang X. P., Nie H. M., Zheng S. W., Zhang T. L., Zhou D., et al. . (2019). Introduction of exogenous glycolate catabolic pathway can strongly enhances photosynthesis and biomass yield of cucumber grown in a low-CO2 environment. Front. Plant Sci. 10, e702. doi: 10.3389/fpls.2019.00702 - DOI - PMC - PubMed
-
- Dąbrowska-Bronk J., Komar D. N., Rusaczonek A., Kozłowska-Makulska A., Szechyńska-Hebda M., Karpiński S. (2016). β-carbonic anhydrases and carbonic ions uptake positively influence Arabidopsis photosynthesis, oxidative stress tolerance and growth in light dependent manner. J. Plant Physiol. 203, 44–54. doi: 10.1016/j.jplph.2016.05.013 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

