Combined analysis of lipidomics and transcriptomics revealed the key pathways and genes of lipids in light-sensitive albino tea plant (Camellia sinensis cv. Baijiguan)
- PMID: 36330254
- PMCID: PMC9623167
- DOI: 10.3389/fpls.2022.1035119
Combined analysis of lipidomics and transcriptomics revealed the key pathways and genes of lipids in light-sensitive albino tea plant (Camellia sinensis cv. Baijiguan)
Abstract
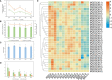
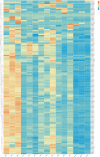
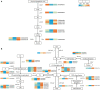
Currently, the mechanism by which light-sensitive albino tea plants respond to light to regulate pigment synthesis has been only partially elucidated. However, few studies have focused on the role of lipid metabolism in the whitening of tea leaves. Therefore, in our study, the leaves of the Baijiguan (BJG) tea tree under shade and light restoration conditions were analyzed by a combination of lipidomics and transcriptomics. The leaf color of BJG was regulated by light intensity and responded to light changes in light by altering the contents and proportions of lipids. According to the correlation analysis, we found three key lipid components that were significantly associated with the chlorophyll SPAD value, namely, MGDG (36:6), DGDG (36:6) and DGDG (34:3). Further weighted gene coexpression network analysis (WGCNA) showed that HY5 TF and GLIP genes may be hub genes involved lipid regulation in albino tea leaves. Our results lay a foundation for further exploration of the color changes in albino tea leaves.
Keywords: Camellia sinensis; WGCNA; albino tea plant; light; lipidomics; transcriptomics.
Copyright © 2022 Zhou, Chen, Wu, Zeng, Chen and Sun.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Block M. A., Dorne A.-J., Joyard J., Douce R. (1983). Preparation and characterization of membrane fractions enriched in outer and inner envelope membranes from spinach chloroplasts. II. biochemical characterization. J. Biol. Chem. 258 (21), 13281–13286. doi: 10.1016/S0021-9258(17)44113-5 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

