Identification of the genes at S and Z reveals the molecular basis and evolution of grass self-incompatibility
- PMID: 36330270
- PMCID: PMC9623065
- DOI: 10.3389/fpls.2022.1011299
Identification of the genes at S and Z reveals the molecular basis and evolution of grass self-incompatibility
Abstract
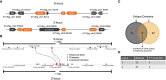
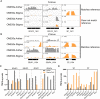
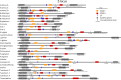
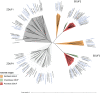
Self-incompatibility (SI) is a feature of many flowering plants, whereby self-pollen is recognized and rejected by the stigma. In grasses (Poaceae), the genes controlling this phenomenon have not been fully elucidated. Grasses have a unique two-locus system, in which two independent genetic loci (S and Z) control self-recognition. S and Z are thought to have arisen from an ancient duplication, common to all grasses. With new chromosome-scale genome data, we examined the genes present at S- and Z-loci, firstly in ryegrass (Lolium perenne), and subsequently in ~20 other grass species. We found that two DUF247 genes and a short unstructured protein (SP/ZP) were present at both S- and Z- in all SI species, while in self-compatible species these genes were often lost or mutated. Expression data suggested that DUF247 genes acted as the male components and SP/ZP were the female components. Consistent with their role in distinguishing self- from non-self, all genes were hypervariable, although key secondary structure features were conserved, including the predicted N-terminal cleavage site of SP/ZP. The evolutionary history of these genes was probed, revealing that specificity groups at the Z-locus arose before the advent of various grass subfamilies/species, while specificity groups at the S-locus arose after the split of Panicoideae, Chloridoideae, Oryzoideae and Pooideae. Finally, we propose a model explaining how the proteins encoded at the S and Z loci might function to specify self-incompatibility.
Keywords: DUF247; Poaceae; grass; pollen; reproduction; self-incompatibility; stigma.
Copyright © 2022 Herridge, McCourt, Jacobs, Mace, Brownfield and Macknight.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Baumann U. (2000). Self-incompatibility in the grasses. Ann. Bot. 85, 203–209. doi: 10.1006/anbo.1999.1056 - DOI
LinkOut - more resources
Full Text Sources

