CRISPR-Cas9 Expressed in Stably Transduced Cell Lines Promotes Recombination and Selects for Herpes Simplex Virus Recombinants
- PMID: 36330462
- PMCID: PMC9629518
- DOI: 10.1016/j.crviro.2022.100023
CRISPR-Cas9 Expressed in Stably Transduced Cell Lines Promotes Recombination and Selects for Herpes Simplex Virus Recombinants
Abstract
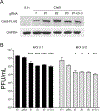
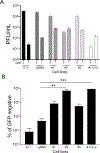
Recombinant herpes simplex virus strains can be constructed by several methods, including homologous recombination, bacterial artificial chromosome manipulation, and yeast genetic methods. Homologous recombination may have the advantage of introducing fewer genetic alterations in the viral genome, but the low level of recombinants can make this method more time consuming if there is no screen or selection. In this study we used complementing cell lines that express Cas9 and guide RNAs targeting the parental virus to rapidly generate recombinant viruses. Analysis of the progeny viruses indicated that CRISPR-Cas9 both promoted recombination to increase recombinant viruses and selected against parental viruses in the transfection progeny viruses. This approach can also be used to enrich for recombinants made by any of the current methods.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
