HIV specific CD8+ TRM-like cells in tonsils express exhaustive signatures in the absence of natural HIV control
- PMID: 36330531
- PMCID: PMC9623418
- DOI: 10.3389/fimmu.2022.912038
HIV specific CD8+ TRM-like cells in tonsils express exhaustive signatures in the absence of natural HIV control
Abstract
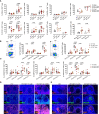
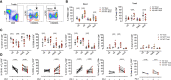
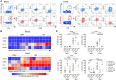
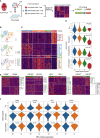
Lymphoid tissues are an important HIV reservoir site that persists in the face of antiretroviral therapy and natural immunity. Targeting these reservoirs by harnessing the antiviral activity of local tissue-resident memory (TRM) CD8+ T-cells is of great interest, but limited data exist on TRM-like cells within lymph nodes of people living with HIV (PLWH). Here, we studied tonsil CD8+ T-cells obtained from PLWH and uninfected controls from South Africa. We show that these cells are preferentially located outside the germinal centers (GCs), the main reservoir site for HIV, and display a low cytolytic and a transcriptionally TRM-like profile distinct from blood CD8+ T-cells. In PLWH, CD8+ TRM-like cells are expanded and adopt a more cytolytic, activated, and exhausted phenotype not reversed by antiretroviral therapy (ART). This phenotype was enhanced in HIV-specific CD8+ T-cells from tonsils compared to matched blood suggesting a higher antigen burden in tonsils. Single-cell transcriptional and clonotype resolution showed that these HIV-specific CD8+ T-cells in the tonsils express heterogeneous signatures of T-cell activation, clonal expansion, and exhaustion ex-vivo. Interestingly, this signature was absent in a natural HIV controller, who expressed lower PD-1 and CXCR5 levels and reduced transcriptional evidence of T-cell activation, exhaustion, and cytolytic activity. These data provide important insights into lymphoid tissue-derived HIV-specific CD8+ TRM-like phenotypes in settings of HIV remission and highlight their potential for immunotherapy and targeting of the HIV reservoirs.
Keywords: CD8+ TRM cells; HIV; PD-1; exhaustion; natural HIV control; tonsils.
Copyright © 2022 Fardoos, Nyquist, Asowata, Kazer, Singh, Ngoepe, Giandhari, Mthabela, Ramjit, Singh, Karim, Buus, Anderson, Porterfield, Sibiya, Bipath, Moodley, Kuhn, Berger, Nguyen, de Oliveira, Ndung’u, Goulder, Shalek, Leslie and Kløverpris.
Conflict of interest statement
AKS reports compensation for consulting and/or SAB membership from Merck, Honeycomb Biotechnologies, Clarity, Repertoire Immune Medicines, Ochre Bio, Third Rock Ventures, Hovione, Relation Therapeutics, FL82, Empress Therapeutics, and Dahlia Biosciences. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Connick E, Folkvord JM, Lind KT, Rakasz EG, Miles B, Wilson NA, et al. . Compartmentalization of simian immunodeficiency virus replication within secondary lymphoid tissues of rhesus macaques is linked to disease stage and inversely related to localization of virus-specific CTL. J Immunol (2014) 193(11):5613. doi: 10.4049/jimmunol.1401161 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

