LncRNA CCTT-mediated RNA-DNA and RNA-protein interactions facilitate the recruitment of CENP-C to centromeric DNA during kinetochore assembly
- PMID: 36332605
- PMCID: PMC9648614
- DOI: 10.1016/j.molcel.2022.09.022
LncRNA CCTT-mediated RNA-DNA and RNA-protein interactions facilitate the recruitment of CENP-C to centromeric DNA during kinetochore assembly
Abstract
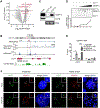
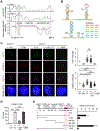
Kinetochore assembly on centromeres is central for chromosome segregation, and defects in this process cause mitotic errors and aneuploidy. Besides the well-established protein network, emerging evidence suggests the involvement of regulatory RNA in kinetochore assembly; however, it has remained elusive about the identity of such RNA, let alone its mechanism of action in this critical process. Here, we report CCTT, a previously uncharacterized long non-coding RNA (lncRNA) transcribed from the arm of human chromosome 17, which plays a vital role in kinetochore assembly. We show that CCTT highly localizes to all centromeres via the formation of RNA-DNA triplex and specifically interacts with CENP-C to help engage this blueprint protein in centromeres, and consequently, CCTT loss triggers extensive mitotic errors and aneuploidy. These findings uncover a non-centromere-derived lncRNA that recruits CENP-C to centromeres and shed critical lights on the function of centromeric DNA sequences as anchor points for kinetochore assembly.
Keywords: CENP-C recruitment; aneuploidy; centromere; lncRNA; mitotic defects.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Similar articles
-
CENP-A Is Dispensable for Mitotic Centromere Function after Initial Centromere/Kinetochore Assembly.Cell Rep. 2016 Nov 22;17(9):2394-2404. doi: 10.1016/j.celrep.2016.10.084. Cell Rep. 2016. PMID: 27880912 Free PMC article.
-
Centromere/kinetochore is assembled through CENP-C oligomerization.Mol Cell. 2023 Jul 6;83(13):2188-2205.e13. doi: 10.1016/j.molcel.2023.05.023. Epub 2023 Jun 8. Mol Cell. 2023. PMID: 37295434
-
Assembly principles and stoichiometry of a complete human kinetochore module.Sci Adv. 2021 Jun 30;7(27):eabg1037. doi: 10.1126/sciadv.abg1037. Print 2021 Jun. Sci Adv. 2021. PMID: 34193424 Free PMC article.
-
Recent insights into mechanisms preventing ectopic centromere formation.Open Biol. 2021 Sep;11(9):210189. doi: 10.1098/rsob.210189. Epub 2021 Sep 8. Open Biol. 2021. PMID: 34493071 Free PMC article. Review.
-
Meiotic CENP-C is a shepherd: bridging the space between the centromere and the kinetochore in time and space.Essays Biochem. 2020 Sep 4;64(2):251-261. doi: 10.1042/EBC20190080. Essays Biochem. 2020. PMID: 32794572 Free PMC article. Review.
Cited by
-
The impact of Yiwei decoction on the LncRNA and CircRNA regulatory networks in premature ovarian insufficiency.Heliyon. 2023 Sep 9;9(9):e20022. doi: 10.1016/j.heliyon.2023.e20022. eCollection 2023 Sep. Heliyon. 2023. PMID: 37809621 Free PMC article.
-
Noncoding RNA-chromatin association: Functions and mechanisms.Fundam Res. 2023 Apr 7;3(5):665-675. doi: 10.1016/j.fmre.2023.03.006. eCollection 2023 Sep. Fundam Res. 2023. PMID: 38933302 Free PMC article. Review.
-
Plant kinetochore complex: composition, function, and regulation.Front Plant Sci. 2024 Oct 10;15:1467236. doi: 10.3389/fpls.2024.1467236. eCollection 2024. Front Plant Sci. 2024. PMID: 39464281 Free PMC article. Review.
-
A New Understanding of Long Non-Coding RNA in Hepatocellular Carcinoma-From m6A Modification to Blood Biomarkers.Cells. 2023 Sep 14;12(18):2272. doi: 10.3390/cells12182272. Cells. 2023. PMID: 37759495 Free PMC article. Review.
-
MLL methyltransferases regulate H3K4 methylation to ensure CENP-A assembly at human centromeres.PLoS Biol. 2023 Jun 28;21(6):e3002161. doi: 10.1371/journal.pbio.3002161. eCollection 2023 Jun. PLoS Biol. 2023. PMID: 37379335 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

