Pathogenic variants in SLF2 and SMC5 cause segmented chromosomes and mosaic variegated hyperploidy
- PMID: 36333305
- PMCID: PMC9636423
- DOI: 10.1038/s41467-022-34349-8
Pathogenic variants in SLF2 and SMC5 cause segmented chromosomes and mosaic variegated hyperploidy
Abstract
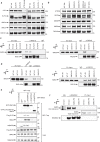
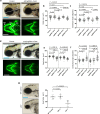
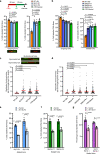
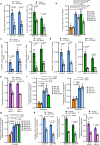
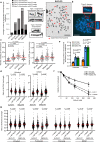
Embryonic development is dictated by tight regulation of DNA replication, cell division and differentiation. Mutations in DNA repair and replication genes disrupt this equilibrium, giving rise to neurodevelopmental disease characterized by microcephaly, short stature and chromosomal breakage. Here, we identify biallelic variants in two components of the RAD18-SLF1/2-SMC5/6 genome stability pathway, SLF2 and SMC5, in 11 patients with microcephaly, short stature, cardiac abnormalities and anemia. Patient-derived cells exhibit a unique chromosomal instability phenotype consisting of segmented and dicentric chromosomes with mosaic variegated hyperploidy. To signify the importance of these segmented chromosomes, we have named this disorder Atelís (meaning - incomplete) Syndrome. Analysis of Atelís Syndrome cells reveals elevated levels of replication stress, partly due to a reduced ability to replicate through G-quadruplex DNA structures, and also loss of sister chromatid cohesion. Together, these data strengthen the functional link between SLF2 and the SMC5/6 complex, highlighting a distinct role for this pathway in maintaining genome stability.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Alcantara D, O’Driscoll M. Congenital microcephaly. Am. J. Med. Genet C. Semin. Med. Genet. 2014;166C:124–139. - PubMed
-
- O’Driscoll M. The pathological consequences of impaired genome integrity in humans; disorders of the DNA replication machinery. J. Pathol. 2017;241:192–207. - PubMed
-
- Grand, R. J. A. & Reynolds, J.J. (eds.). DNA Repair and Replication: Mechanisms and Clinical Significance, 348 (Garland Science, 2018).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

