Intrinsic host susceptibility among multiple species to intranasal SARS-CoV-2 identifies diverse virological, biodistribution and pathological outcomes
- PMID: 36333445
- PMCID: PMC9636276
- DOI: 10.1038/s41598-022-23339-x
Intrinsic host susceptibility among multiple species to intranasal SARS-CoV-2 identifies diverse virological, biodistribution and pathological outcomes
Abstract
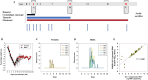
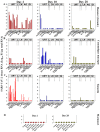
SARS-CoV-2 exhibits a diverse host species range with variable outcomes, enabling differential host susceptibility studies to assess suitability for pre-clinical countermeasure and pathogenesis studies. Baseline virological, molecular and pathological outcomes were determined among multiple species-one Old World non-human primate (NHP) species (cynomolgus macaques), two New World NHP species (red-bellied tamarins; common marmosets) and Syrian hamsters-following single-dose, atraumatic intranasal administration of SARS-CoV-2/Victoria-01. After serial sacrifice 2, 10 and 28-days post-infection (dpi), hamsters and cynomolgus macaques displayed differential virus biodistribution across respiratory, gastrointestinal and cardiovascular systems. Uniquely, New World tamarins, unlike marmosets, exhibited high levels of acute upper airway infection, infectious virus recovery associated with mild lung pathology representing a host previously unrecognized as susceptible to SARS-CoV-2. Across all species, lung pathology was identified post-clearance of virus shedding (antigen/RNA), with an association of virus particles within replication organelles in lung sections analysed by electron microscopy. Disrupted cell ultrastructure and lung architecture, including abnormal morphology of mitochondria 10-28 dpi, represented on-going pathophysiological consequences of SARS-CoV-2 in predominantly asymptomatic hosts. Infection kinetics and host pathology comparators using standardized methodologies enables model selection to bridge differential outcomes within upper and lower respiratory tracts and elucidate longer-term consequences of asymptomatic SARS-CoV-2 infection.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Muñoz-Fontela C, Dowling WE, Funnell SGP, Gsell PS, Riveros-Balta AX, Albrecht RA, Andersen H, Baric RS, Carroll MW, Cavaleri M, Qin C, Crozier I, Dallmeier K, de Waal L, de Wit E, Delang L, Dohm E, Duprex WP, Falzarano D, Finch CL, Frieman MB, Graham BS, Gralinski LE, Guilfoyle K, Haagmans BL, Hamilton GA, Hartman AL, Herfst S, Kaptein SJF, Klimstra WB, Knezevic I, Krause PR, Kuhn JH, Le Grand R, Lewis MG, Liu WC, Maisonnasse P, McElroy AK, Munster V, Oreshkova N, Rasmussen AL, Rocha-Pereira J, Rockx B, Rodríguez E, Rogers TF, Salguero FJ, Schotsaert M, Stittelaar KJ, Thibaut HJ, Tseng CT, Vergara-Alert J, Beer M, Brasel T, Chan JFW, García-Sastre A, Neyts J, Perlman S, Reed DS, Richt JA, Roy CJ, Segalés J, Vasan SS, Henao-Restrepo AM, Barouch DH. Animal models for COVID-19. Nature. 2020;586(7830):509–515. - PMC - PubMed
-
- Damas J, Hughes GM, Keough KC, Painter CA, Persky NS, Corbo M, Hiller M, Koepfli KP, Pfenning AR, Zhao H, Genereux DP, Swofford R, Pollard KS, Ryder OA, Nweeia MT, Lindblad-Toh K, Teeling EC, Karlsson EK, Lewin HA. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE-2 in vertebrates. Proc. Natl. Acad. Sci. USA. 2020;117(36):22311–22322. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

