Whole-genome sequencing of chronic lymphocytic leukemia identifies subgroups with distinct biological and clinical features
- PMID: 36333502
- PMCID: PMC9649442
- DOI: 10.1038/s41588-022-01211-y
Whole-genome sequencing of chronic lymphocytic leukemia identifies subgroups with distinct biological and clinical features
Abstract
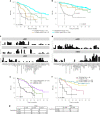
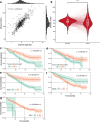
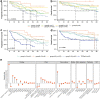
The value of genome-wide over targeted driver analyses for predicting clinical outcomes of cancer patients is debated. Here, we report the whole-genome sequencing of 485 chronic lymphocytic leukemia patients enrolled in clinical trials as part of the United Kingdom's 100,000 Genomes Project. We identify an extended catalog of recurrent coding and noncoding genetic mutations that represents a source for future studies and provide the most complete high-resolution map of structural variants, copy number changes and global genome features including telomere length, mutational signatures and genomic complexity. We demonstrate the relationship of these features with clinical outcome and show that integration of 186 distinct recurrent genomic alterations defines five genomic subgroups that associate with response to therapy, refining conventional outcome prediction. While requiring independent validation, our findings highlight the potential of whole-genome sequencing to inform future risk stratification in chronic lymphocytic leukemia.
© 2022. The Author(s).
Conflict of interest statement
In the past five years, A.S. has received in-kind contributions from Illumina and Oxford Nanopore Technology and is a shareholder of Illumina. She is a company director and shareholder of SERENOx Ltd. A.S. has received honoraria from Exact Sciences, Janssen, Astra Zeneca, Abbvie and Beigene, non-restricted research grants from Janssen and Astra Zeneca and an educational grant from Abbvie. A.R.P. receives research funding from Celgene/BMS, Gilead, Napp and Roche. N.A. received speaker fees from Gilead. P.A., T.J., U.M., M.R. and D.B. are employees of Illumina, a public company that develops and markets systems for genetic analysis. The remaining authors declare no competing interests.
Figures















References
Publication types
MeSH terms
Grants and funding
- C124388/CRUK_/Cancer Research UK/United Kingdom
- C42023/A29370/CRUK_/Cancer Research UK/United Kingdom
- DH_/Department of Health/United Kingdom
- MR/M009203/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_00029/4/MRC_/Medical Research Council/United Kingdom
- MR/R008108/1/MRC_/Medical Research Council/United Kingdom
- C34999/A18087/CRUK_/Cancer Research UK/United Kingdom
- MC_EX_MR/M009203/1/MRC_/Medical Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- MR/N00969X/1/MRC_/Medical Research Council/United Kingdom
- 23669/CRUK_/Cancer Research UK/United Kingdom
- 29370/CRUK_/Cancer Research UK/United Kingdom
- C24563/A15581/CRUK_/Cancer Research UK/United Kingdom
- 12362/CRUK_/Cancer Research UK/United Kingdom
- 214388/WT_/Wellcome Trust/United Kingdom
- C2750/A23669/CRUK_/Cancer Research UK/United Kingdom
- MC_PC_14089/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

