A novel and dual digestive symbiosis scales up the nutrition and immune system of the holobiont Rimicaris exoculata
- PMID: 36333777
- PMCID: PMC9636832
- DOI: 10.1186/s40168-022-01380-2
A novel and dual digestive symbiosis scales up the nutrition and immune system of the holobiont Rimicaris exoculata
Abstract
Background: In deep-sea hydrothermal vent areas, deprived of light, most animals rely on chemosynthetic symbionts for their nutrition. These symbionts may be located on their cuticle, inside modified organs, or in specialized cells. Nonetheless, many of these animals have an open and functional digestive tract. The vent shrimp Rimicaris exoculata is fueled mainly by its gill chamber symbionts, but also has a complete digestive system with symbionts. These are found in the shrimp foregut and midgut, but their roles remain unknown. We used genome-resolved metagenomics on separate foregut and midgut samples, taken from specimens living at three contrasted sites along the Mid-Atlantic Ridge (TAG, Rainbow, and Snake Pit) to reveal their genetic potential.
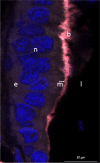
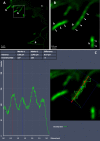
Results: We reconstructed and studied 20 Metagenome-Assembled Genomes (MAGs), including novel lineages of Hepatoplasmataceae and Deferribacteres, abundant in the shrimp foregut and midgut, respectively. Although the former showed streamlined reduced genomes capable of using mostly broken-down complex molecules, Deferribacteres showed the ability to degrade complex polymers, synthesize vitamins, and encode numerous flagellar and chemotaxis genes for host-symbiont sensing. Both symbionts harbor a diverse set of immune system genes favoring holobiont defense. In addition, Deferribacteres were observed to particularly colonize the bacteria-free ectoperitrophic space, in direct contact with the host, elongating but not dividing despite possessing the complete genetic machinery necessary for this.
Conclusion: Overall, these data suggest that these digestive symbionts have key communication and defense roles, which contribute to the overall fitness of the Rimicaris holobiont. Video Abstract.
Keywords: Deferribacteres; Digestive symbiosis; Hepatoplasmataceae; Immunity; Metagenomics; Rimicaris exoculata.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- McFall-Ngai MJ. Unseen forces: the influence of bacteria on animal development. Dev Biol. 2002;242:1–14. - PubMed
-
- Zilber-Rosenberg I, Rosenberg E. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev. 2008;32:723–735. - PubMed
-
- Dubilier N, Bergin C, Lott C. Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat Rev Microbiol. 2008;6:725–740. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

