Androgen deprivation restores ARHGEF2 to promote neuroendocrine differentiation of prostate cancer
- PMID: 36335093
- PMCID: PMC9637107
- DOI: 10.1038/s41419-022-05366-8
Androgen deprivation restores ARHGEF2 to promote neuroendocrine differentiation of prostate cancer
Erratum in
-
Correction: Androgen deprivation restores ARHGEF2 to promote neuroendocrine differentiation of prostate cancer.Cell Death Dis. 2023 Oct 2;14(10):647. doi: 10.1038/s41419-023-06051-0. Cell Death Dis. 2023. PMID: 37783705 Free PMC article. No abstract available.
Abstract
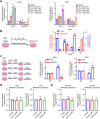
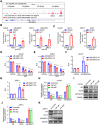
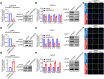
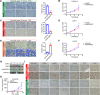
Androgen receptor (AR) plays an important role in the progression of prostate cancer and has been targeted by castration or AR-antagonists. The emergence of castration-resistant prostate cancer (CRPC) after androgen deprivation therapy (ADT) is inevitable. However, it is not entirely clear how ADT fails or how it causes resistance. Through analysis of RNA-seq data, we nominate ARHGEF2 as a pivotal androgen-repressed gene. We show that ARHGEF2 is directly suppressed by androgen/AR. AR occupies the enhancer and communicates with the promoter region of ARHGEF2. Functionally, ARHGEF2 is important for the growth, lethal phenotype, and survival of CRPC cells and tumor xenografts. Correspondingly, AR inhibition or AR antagonist treatment can restore ARHGEF2 expression, thereby allowing prostate cancer cells to induce treatment resistance and tolerance. Overall, our findings provide an explanation for the contradictory clinical results that ADT resistance may be caused by the up-regulation of ARHGEF2 and provide a novel target.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

