Analysis of clinically relevant variants from ancestrally diverse Asian genomes
- PMID: 36335097
- PMCID: PMC9637116
- DOI: 10.1038/s41467-022-34116-9
Analysis of clinically relevant variants from ancestrally diverse Asian genomes
Abstract
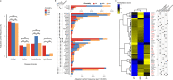
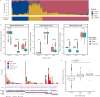
Asian populations are under-represented in human genomics research. Here, we characterize clinically significant genetic variation in 9051 genomes representing East Asian, South Asian, and severely under-represented Austronesian-speaking Southeast Asian ancestries. We observe disparate genetic risk burden attributable to ancestry-specific recurrent variants and identify individuals with variants specific to ancestries discordant to their self-reported ethnicity, mostly due to cryptic admixture. About 27% of severe recessive disorder genes with appreciable carrier frequencies in Asians are missed by carrier screening panels, and we estimate 0.5% Asian couples at-risk of having an affected child. Prevalence of medically-actionable variant carriers is 3.4% and a further 1.6% harbour variants with potential for pathogenic classification upon additional clinical/experimental evidence. We profile 23 pharmacogenes with high-confidence gene-drug associations and find 22.4% of Asians at-risk of Centers for Disease Control and Prevention Tier 1 genetic conditions concurrently harbour pharmacogenetic variants with actionable phenotypes, highlighting the benefits of pre-emptive pharmacogenomics. Our findings illuminate the diversity in genetic disease epidemiology and opportunities for precision medicine for a large, diverse Asian population.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Popejoy AB. Too many scientists still say Caucasian. Nature. 2021;596:463.
-
- Precision medicine needs an equity agenda. Nat. Med. 27, 737. 10.1038/s41591-021-01373-y (2021). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

