Label-free third harmonic generation imaging and quantification of lipid droplets in live filamentous fungi
- PMID: 36335164
- PMCID: PMC9637149
- DOI: 10.1038/s41598-022-23502-4
Label-free third harmonic generation imaging and quantification of lipid droplets in live filamentous fungi
Abstract
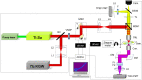
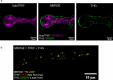
We report the utilization of Third-Harmonic Generation microscopy for label-free live cell imaging of lipid droplets in the hypha of filamentous fungus Phycomyces blakesleeanus. THG microscopy images showed bright spherical features dispersed throughout the hypha cytoplasm in control conditions and a transient increase in the number of bright features after complete nitrogen starvation. Colocalization analysis of THG and lipid-counterstained images disclosed that the cytoplasmic particles were lipid droplets. Particle Size Analysis and Image Correlation Spectroscopy were used to quantify the number density and size of lipid droplets. The two analysis methods both revealed an increase from 16 × 10-3 to 23 × 10-3 lipid droplets/µm2 after nitrogen starvation and a decrease in the average size of the droplets (range: 0.5-0.8 µm diameter). In conclusion, THG imaging, followed by PSA and ICS, can be reliably used for filamentous fungi for the in vivo quantification of lipid droplets without the need for labeling and/or fixation. In addition, it has been demonstrated that ICS is suitable for THG microscopy.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Distinction between breast cancer cell subtypes using third harmonic generation microscopy.J Biophotonics. 2017 Sep;10(9):1152-1162. doi: 10.1002/jbio.201600173. Epub 2016 Oct 18. J Biophotonics. 2017. PMID: 27753229
-
Third Harmonic Generation microscopy distinguishes malignant cell grade in human breast tissue biopsies.Sci Rep. 2020 Jul 6;10(1):11055. doi: 10.1038/s41598-020-67857-y. Sci Rep. 2020. PMID: 32632110 Free PMC article.
-
Imaging Cytometry of Human Leukocytes with Third Harmonic Generation Microscopy.Sci Rep. 2016 Nov 15;6:37210. doi: 10.1038/srep37210. Sci Rep. 2016. PMID: 27845443 Free PMC article.
-
Third harmonic generation microscopy of cells and tissue organization.J Cell Sci. 2016 Jan 15;129(2):245-55. doi: 10.1242/jcs.152272. Epub 2016 Jan 7. J Cell Sci. 2016. PMID: 26743082 Review.
-
Gravisusception by buoyancy: a mechanism ubiquitous among fungi?Protoplasma. 2006 Dec;229(2-4):117-23. doi: 10.1007/s00709-006-0218-7. Epub 2006 Dec 16. Protoplasma. 2006. PMID: 17180492 Review.
Cited by
-
ATP modulation of osmotically activated anionic current in the membrane of Phycomyces blakesleeanus sporangiophore.Sci Rep. 2023 Jul 24;13(1):11897. doi: 10.1038/s41598-023-39021-9. Sci Rep. 2023. PMID: 37488205 Free PMC article.
References
-
- Weigelin B, Bakker GJ, Friedl P. Third harmonic generation microscopy of cells and tissue organization. J. Cell Sci. 2016;129:245–255. - PubMed
-
- Yelin D, Silberberg Y. Laser scanning third-harmonic-generation microscopy in biology. Opt. Express. 1999;5(8):169–175. - PubMed
-
- Barzda V, et al. Visualization of mitochondria in cardiomyocytes. Opt. Express. 2005;13:8263. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

