Generative adversarial network-created brain SPECTs of cerebral ischemia are indistinguishable to scans from real patients
- PMID: 36335166
- PMCID: PMC9637159
- DOI: 10.1038/s41598-022-23325-3
Generative adversarial network-created brain SPECTs of cerebral ischemia are indistinguishable to scans from real patients
Abstract
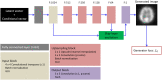
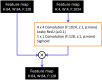
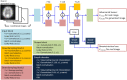
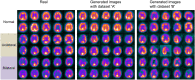
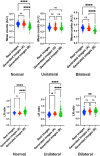
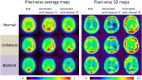
Deep convolutional generative adversarial networks (GAN) allow for creating images from existing databases. We applied a modified light-weight GAN (FastGAN) algorithm to cerebral blood flow SPECTs and aimed to evaluate whether this technology can generate created images close to real patients. Investigating three anatomical levels (cerebellum, CER; basal ganglia, BG; cortex, COR), 551 normal (248 CER, 174 BG, 129 COR) and 387 pathological brain SPECTs using N-isopropyl p-I-123-iodoamphetamine (123I-IMP) were included. For the latter scans, cerebral ischemic disease comprised 291 uni- (66 CER, 116 BG, 109 COR) and 96 bilateral defect patterns (44 BG, 52 COR). Our model was trained using a three-compartment anatomical input (dataset 'A'; including CER, BG, and COR), while for dataset 'B', only one anatomical region (COR) was included. Quantitative analyses provided mean counts (MC) and left/right (LR) hemisphere ratios, which were then compared to quantification from real images. For MC, 'B' was significantly different for normal and bilateral defect patterns (P < 0.0001, respectively), but not for unilateral ischemia (P = 0.77). Comparable results were recorded for LR, as normal and ischemia scans were significantly different relative to images acquired from real patients (P ≤ 0.01, respectively). Images provided by 'A', however, revealed comparable quantitative results when compared to real images, including normal (P = 0.8) and pathological scans (unilateral, P = 0.99; bilateral, P = 0.68) for MC. For LR, only uni- (P = 0.03), but not normal or bilateral defect scans (P ≥ 0.08) reached significance relative to images of real patients. With a minimum of only three anatomical compartments serving as stimuli, created cerebral SPECTs are indistinguishable to images from real patients. The applied FastGAN algorithm may allow to provide sufficient scan numbers in various clinical scenarios, e.g., for "data-hungry" deep learning technologies or in the context of orphan diseases.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Goodfellow, I.J., Pouget-Abadie, J., Mirza, M., Xu, B., Warde-Farley, D., Ozair, S., et al. Generative Adversarial Networks. ArXiv e-prints [Internet]. 2014 June 01, 2014. https://ui.adsabs.harvard.edu/#abs/2014arXiv1406.2661G.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

