Analysis of the twenty-six largest outbreaks of tuberculosis in Aragon using whole-genome sequencing for surveillance purposes
- PMID: 36335223
- PMCID: PMC9637126
- DOI: 10.1038/s41598-022-23343-1
Analysis of the twenty-six largest outbreaks of tuberculosis in Aragon using whole-genome sequencing for surveillance purposes
Abstract
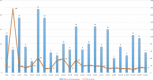
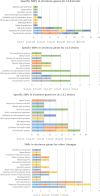
The incidence of tuberculosis in Aragon, Spain, is around ten cases per 100,000 inhabitants. Since 2004, a molecular surveillance protocol has been carried out; therefore, all M. tuberculosis strains are genotyped. Recently, whole-genome sequencing has been implemented for relevant isolates. The aim of this work is to characterise at the molecular level the causative strains of the 26 largest outbreaks of the community (including ten or more cases), genotyped by IS6110-RFLP and causing 26% of tuberculosis cases. To achieve this objective, two or three isolates of each IS6110-cluster belonging to different years were selected for sequencing. We found that strains of lineages L4.8, L4.3 and L4.1.2 were the most frequent. The threshold of 12 SNPs as the maximum distance for confirming the belonging to an outbreak was met for 18 of the 26 IS6110-clusters. Four pairs of isolates with more than 90 SNPs were identified as not belonging to the same strain, and four other pairs were kept in doubt as the number of SNPs was close to 12, between 14 and 35. The study of Regions of Difference revealed that they are lineage conserved. Moreover, we could analyse the IS6110 locations for all genome-sequenced isolates, finding some frequent locations in isolates belonging to the same lineage and certain IS6110 movements between the paired isolates. In the vast majority, these movements were not captured by the IS6110-RFLP pattern. After classifying the genes containing SNP by their functional category, we could confirm that the number of SNPs detected in genes considered as virulence factors and the number of cases the strain produced were not related, suggesting that a particular SNP is more relevant than the number. The characteristics found in the most successful strains in our community could be useful for other researchers in epidemiology, virulence and pathogenesis.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
The MtZ Strain: Molecular Characteristics and Outbreak Investigation of the Most Successful Mycobacterium tuberculosis Strain in Aragon Using Whole-Genome Sequencing.Front Cell Infect Microbiol. 2022 May 24;12:887134. doi: 10.3389/fcimb.2022.887134. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35685752 Free PMC article.
-
[Future prospects of molecular epidemiology in tuberculosis].Kekkaku. 2009 Dec;84(12):783-4. Kekkaku. 2009. PMID: 20077862 Japanese.
-
IS6110-RFLP and spoligotyping of Mycobacterium tuberculosis isolates in Iran.Scand J Infect Dis. 2000;32(6):663-8. doi: 10.1080/003655400459595. Scand J Infect Dis. 2000. PMID: 11200378
-
A whole-genome sequencing study of an X-family tuberculosis outbreak focus on transmission chain along 25 years.Tuberculosis (Edinb). 2021 Jan;126:102022. doi: 10.1016/j.tube.2020.102022. Epub 2020 Nov 28. Tuberculosis (Edinb). 2021. PMID: 33341027 Review.
-
[Molecular epidemiology of Mycobacterium tuberculosis using by RFLP analysis between genomic DNA--its accomplishment and practice].Kekkaku. 2003 Oct;78(10):641-51. Kekkaku. 2003. PMID: 14621573 Review. Japanese.
Cited by
-
Interred mechanisms of resistance and host immune evasion revealed through network-connectivity analysis of M. tuberculosis complex graph pangenome.mSystems. 2025 Apr 22;10(4):e0049924. doi: 10.1128/msystems.00499-24. Epub 2025 Mar 6. mSystems. 2025. PMID: 40261029 Free PMC article.
-
Genetic diversity and transmission pattern of multidrug-resistant tuberculosis based on whole-genome sequencing in Wuhan, China.Front Public Health. 2025 Jan 28;13:1442987. doi: 10.3389/fpubh.2025.1442987. eCollection 2025. Front Public Health. 2025. PMID: 39935875 Free PMC article.
References
-
- WHO. WHO | Global tuberculosis report 2019. Geneva: World Health Organization. Licence: CC BY-NC-SA 3.0 IGO (2019).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

