Leukemia inhibitory factor is a therapeutic target for renal interstitial fibrosis
- PMID: 36335669
- PMCID: PMC9646860
- DOI: 10.1016/j.ebiom.2022.104312
Leukemia inhibitory factor is a therapeutic target for renal interstitial fibrosis
Abstract
Background: The role of the IL6 family members in organ fibrosis, including renal interstitial fibrosis (TIF), has been widely explored. However, few studies have ever simultaneously examined them in the same cohort of patients. Besides, the role of leukemia inhibitory factor (LIF) in TIF remains unclear.
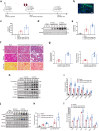
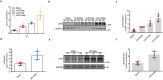
Methods: RNA-seq data of kidney biopsies from chronic kidney disease (CKD) patients, in both public databases and our assays, were used to analyze transcript levels of IL6 family members. Two TIF mouse models, the unilateral ureteral obstruction (UUO) and the ischemia reperfusion injury (IRI), were employed to validate the finding. To assess the role of LIF in vivo, short hairpin RNA, lenti-GFP-LIF was used to knockdown LIF receptor (LIFR), overexpress LIF, respectively. LIF-neutralizing antibody was used in therapeutic studies. Whether urinary LIF could be used as a promising predictor for CKD progression was investigated in a prospective observation patient cohort.
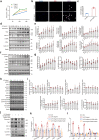
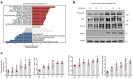
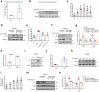
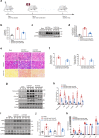
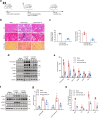
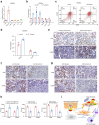
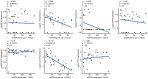
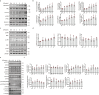
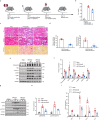
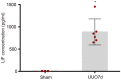
Findings: Among IL6 family members, LIF is the most upregulated one in both human and mouse renal fibrotic lesions. The mRNA level of LIF negatively correlated with eGFR with the strongest correlation and the smallest P value. Baseline urinary concentrations of LIF in CKD patients predict the risk of CKD progression to end-stage kidney disease by Kaplan-Meier analysis. In mouse TIF models, knockdown of LIFR alleviated TIF; conversely, overexpressing LIF exacerbated TIF. Most encouragingly, visible efficacy against TIF was observed by administering LIF-neutralizing antibodies to mice. Mechanistically, LIF-LIFR-EGR1 axis and Sonic Hedgehog signaling formed a vicious cycle between fibroblasts and proximal tubular cells to augment LIF expression and promote the pro-fibrotic response via ERK and STAT3 activation.
Interpretation: This study discovered that LIF is a noninvasive biomarker for the progression of CKD and a potential therapeutic target of TIF.
Fundings: Stated in the Acknowledgements section of the manuscript.
Keywords: Chronic kidney disease; Fibroblast activation; IL6 family; LIF; Renal fibrosis.
Copyright © 2022 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests All the authors declared no conflict of interest.
Figures







Comment in
-
Tubule epithelial cells and fibroblasts communication: Vicious cycle of renal fibrosis.EBioMedicine. 2022 Dec;86:104360. doi: 10.1016/j.ebiom.2022.104360. Epub 2022 Nov 10. EBioMedicine. 2022. PMID: 36371984 Free PMC article. No abstract available.
Similar articles
-
The ILEI/LIFR complex induces EMT via the Akt and ERK pathways in renal interstitial fibrosis.J Transl Med. 2022 Jan 29;20(1):54. doi: 10.1186/s12967-022-03265-2. J Transl Med. 2022. PMID: 35093095 Free PMC article.
-
Leukemia inhibitory factor is involved in tubular regeneration after experimental acute renal failure.J Am Soc Nephrol. 2003 Dec;14(12):3090-101. doi: 10.1097/01.asn.0000101180.96787.02. J Am Soc Nephrol. 2003. PMID: 14638908
-
Leukemia inhibitory factor functions in parallel with interleukin-6 to promote ovarian cancer growth.Oncogene. 2019 Feb;38(9):1576-1584. doi: 10.1038/s41388-018-0523-6. Epub 2018 Oct 10. Oncogene. 2019. PMID: 30305729 Free PMC article.
-
Renal fibrosis: Primacy of the proximal tubule.Matrix Biol. 2018 Aug;68-69:248-262. doi: 10.1016/j.matbio.2018.02.006. Epub 2018 Feb 6. Matrix Biol. 2018. PMID: 29425694 Free PMC article. Review.
-
Hypoxia and Renal Tubulointerstitial Fibrosis.Adv Exp Med Biol. 2019;1165:467-485. doi: 10.1007/978-981-13-8871-2_23. Adv Exp Med Biol. 2019. PMID: 31399980 Review.
Cited by
-
Identification of novel immune-related signatures for keloid diagnosis and treatment: insights from integrated bulk RNA-seq and scRNA-seq analysis.Hum Genomics. 2024 Jul 16;18(1):80. doi: 10.1186/s40246-024-00647-z. Hum Genomics. 2024. PMID: 39014455 Free PMC article.
-
Connexin-43 hemichannels orchestrate NOD-like receptor protein-3 (NLRP3) inflammasome activation and sterile inflammation in tubular injury.Cell Commun Signal. 2023 Sep 28;21(1):263. doi: 10.1186/s12964-023-01245-7. Cell Commun Signal. 2023. PMID: 37770948 Free PMC article.
-
Knockout of integrin αvβ6 protects against renal inflammation in chronic kidney disease by reduction of pro-inflammatory macrophages.Cell Death Dis. 2024 Jun 6;15(6):397. doi: 10.1038/s41419-024-06785-5. Cell Death Dis. 2024. PMID: 38844455 Free PMC article.
-
Role of Interleukin-6 Family Cytokines in Organ Fibrosis.Kidney Dis (Basel). 2023 Mar 22;9(4):239-253. doi: 10.1159/000530288. eCollection 2023 Aug. Kidney Dis (Basel). 2023. PMID: 37900004 Free PMC article. Review.
-
Role of circulating inflammatory protein in the development of diabetic renal complications: proteome-wide Mendelian randomization and colocalization analyses.Front Endocrinol (Lausanne). 2024 Jul 8;15:1406442. doi: 10.3389/fendo.2024.1406442. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39040677 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

