A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors
- PMID: 36335936
- PMCID: PMC9531661
- DOI: 10.1016/j.cell.2022.09.037
A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors
Abstract
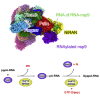
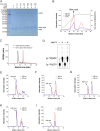
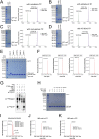
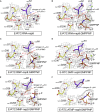
Decoration of cap on viral RNA plays essential roles in SARS-CoV-2 proliferation. Here, we report a mechanism for SARS-CoV-2 RNA capping and document structural details at atomic resolution. The NiRAN domain in polymerase catalyzes the covalent link of RNA 5' end to the first residue of nsp9 (termed as RNAylation), thus being an intermediate to form cap core (GpppA) with GTP catalyzed again by NiRAN. We also reveal that triphosphorylated nucleotide analog inhibitors can be bonded to nsp9 and fit into a previously unknown "Nuc-pocket" in NiRAN, thus inhibiting nsp9 RNAylation and formation of GpppA. S-loop (residues 50-KTN-52) in NiRAN presents a remarkable conformational shift observed in RTC bound with sofosbuvir monophosphate, reasoning an "induce-and-lock" mechanism to design inhibitors. These findings not only improve the understanding of SARS-CoV-2 RNA capping and the mode of action of NAIs but also provide a strategy to design antiviral drugs.
Keywords: SARS-CoV-2; capping; cryo-EM; nucleotide analog inhibitor.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures








Similar articles
-
Conserved Characteristics of NMPylation Activities of Alpha- and Betacoronavirus NiRAN Domains.J Virol. 2023 Jun 29;97(6):e0046523. doi: 10.1128/jvi.00465-23. Epub 2023 May 18. J Virol. 2023. PMID: 37199624 Free PMC article.
-
The mechanism of RNA capping by SARS-CoV-2.Nature. 2022 Sep;609(7928):793-800. doi: 10.1038/s41586-022-05185-z. Epub 2022 Aug 9. Nature. 2022. PMID: 35944563 Free PMC article.
-
Response to: The mechanism for GTP-mediated RNA capping by the SARS-CoV-2 NiRAN domain remains unresolved.Cell. 2025 Aug 7;188(16):4462-4469.e9. doi: 10.1016/j.cell.2025.05.045. Epub 2025 Jun 25. Cell. 2025. PMID: 40570837
-
State-of-the-Art Molecular Dynamics Simulation Studies of RNA-Dependent RNA Polymerase of SARS-CoV-2.Int J Mol Sci. 2022 Sep 8;23(18):10358. doi: 10.3390/ijms231810358. Int J Mol Sci. 2022. PMID: 36142270 Free PMC article. Review.
-
Redefining pseudokinases: A look at the untapped enzymatic potential of pseudokinases.IUBMB Life. 2023 Apr;75(4):370-376. doi: 10.1002/iub.2698. Epub 2023 Jan 5. IUBMB Life. 2023. PMID: 36602414 Free PMC article. Review.
Cited by
-
Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.bioRxiv [Preprint]. 2023 Nov 20:2023.11.17.567629. doi: 10.1101/2023.11.17.567629. bioRxiv. 2023. Update in: Protein Cell. 2024 Jul 1;15(7):547-552. doi: 10.1093/procel/pwae009. PMID: 38045375 Free PMC article. Updated. Preprint.
-
Identification of TMEM53 as a novel SADS-CoV restriction factor that targets viral RNA-dependent RNA polymerase.Emerg Microbes Infect. 2023 Dec;12(2):2249120. doi: 10.1080/22221751.2023.2249120. Emerg Microbes Infect. 2023. PMID: 37584551 Free PMC article.
-
New insights into the pathogenesis of SARS-CoV-2 during and after the COVID-19 pandemic.Front Immunol. 2024 Jun 7;15:1363572. doi: 10.3389/fimmu.2024.1363572. eCollection 2024. Front Immunol. 2024. PMID: 38911850 Free PMC article. Review.
-
SND1 binds SARS-CoV-2 negative-sense RNA and promotes viral RNA synthesis through NSP9.Cell. 2023 Oct 26;186(22):4834-4850.e23. doi: 10.1016/j.cell.2023.09.002. Epub 2023 Oct 3. Cell. 2023. PMID: 37794589 Free PMC article.
-
SHIFTR enables the unbiased identification of proteins bound to specific RNA regions in live cells.Nucleic Acids Res. 2024 Mar 21;52(5):e26. doi: 10.1093/nar/gkae038. Nucleic Acids Res. 2024. PMID: 38281241 Free PMC article.
References
-
- Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

