Computational modeling of the effect of five mutations on the structure of the ACE2 receptor and their correlation with infectivity and virulence of some emerged variants of SARS-CoV-2 suggests mechanisms of binding affinity dysregulation
- PMID: 36336003
- PMCID: PMC9630301
- DOI: 10.1016/j.cbi.2022.110244
Computational modeling of the effect of five mutations on the structure of the ACE2 receptor and their correlation with infectivity and virulence of some emerged variants of SARS-CoV-2 suggests mechanisms of binding affinity dysregulation
Abstract
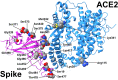
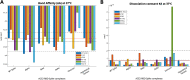
Interactions between the human angiotensin-converting enzyme 2 (ACE2) and the RBD region of the SARS-CoV-2 Spike protein are critical for virus entry into the host cell. The objective of this work was to identify some of the most relevant SARS-CoV-2 Spike variants that emerged during the pandemic and evaluate their binding affinity with human variants of ACE2 since some ACE2 variants can enhance or reduce the affinity of the interaction between the ACE2 and S proteins. However, no information has been sought to extrapolate to different variants of SARS-CoV-2. Therefore, to understand the impact on the affinity of the interaction between ACE2 protein variants and SARS-CoV-2 protein S variants, molecular docking was used in this study to predict the effects of five mutations of ACE2 when they interact with Alpha, Beta, Delta, Omicron variants and a hypothetical variant, which present mutations in the RBD region of the SARS-CoV-2 Spike protein. Our results suggest that these variants could alter the interaction of the Spike and the human ACE2 protein, losing or creating new inter-protein contacts, enhancing viral fitness by improving binding affinity, and leading to an increase in infectivity, virulence, and transmission. This investigation highlighted that the S19P mutation of ACE2 decreases the binding affinity between the ACE2 and Spike proteins in the presence of the Beta variant and the wild-type variant of SARS-CoV-2 isolated in Wuhan-2019. The R115Q mutation of ACE2 lowers the binding affinity of these two proteins in the presence of the Beta and Delta variants. Similarly, the K26R mutation lowers the affinity of the interaction between the ACE2 and Spike proteins in the presence of the Alpha variant. This decrease in binding affinity is probably due to the lack of interaction between some of the key residues of the interaction complex between the ACE2 protein and the RBD region of the SARS-CoV-2 Spike protein. Therefore, ACE2 mutations appear in the presence of these variants, they could suggest an intrinsic resistance to COVID-19 disease. On the other hand, our results suggested that the K26R, M332L, and K341R mutations of ACE2 expressively showed the affinity between the ACE2 and Spike proteins in the Alpha, Beta, and Delta variants. Consequently, these ACE2 mutations in the presence of the Alpha, Beta, and delta variants of SARS-CoV-2 could be more infectious and virulent in human cells compared to the SARS-CoV-2 isolated in Wuhan-2019 and it could have a negative prognosis of the disease. Finally, the Omicron variant in interaction with ACE2 WT, S19P, R115Q, M332L, and K341R mutations of ACE2 showed a significant decrease in binding affinity. This could be consistent that the Omicron variant causes less severe symptoms than previous variants. On the other hand, our results suggested Omicron in the complex with K26R, the binding affinity is increased between ACE2/RBD, which could indicate a negative prognosis of the disease in people with these allelic conditions.
Keywords: ACE2-SARS-CoV-2; Alpha; Beta; Binding affinity; Delta; Omicron.
Copyright © 2022 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
References
-
- Tanaka S., Nelson G., Olson C.A., Buzko O., Higashide W., Shin A., Gonzalez M., Taft J., Patel R., Buta S., Richardson A., Bogunovic D., Spilman P., Niazi K., Rabizadeh S., Soon-Shiong P. An ACE2 Triple Decoy that neutralizes SARS-CoV-2 shows enhanced affinity for virus variants. Sci. Rep. 2021;11(1):1–12. doi: 10.1038/s41598-021-91809-9. - DOI - PMC - PubMed
-
- Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C.C., Muecksch F., Rutkowska M., Hoffmann H.H., Michailidis E., Gaebler C., Agudelo M., Cho A., Wang Z., Gazumyan A., Cipolla M., Luchsinger L., Hillyer C.D., Caskey M.…Bieniasz P.D. Escape from neutralizing antibodies 1 by SARS-CoV-2 spike protein variants. Elife. 2020;9:1. doi: 10.7554/eLife.61312. - DOI - PMC - PubMed
-
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., Oliveira T.Y., Yang Z., Abernathy M.E., Huey-Tubman K.E., Hurley A., Turroja M., West K.A., Gordon K., Millard K.G.…Nussenzweig M.C. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592(7855):616–622. doi: 10.1038/s41586-021-03324-6. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

