Bioinformatic Analysis of B- and T-cell Epitopes from SARS-CoV-2 Structural Proteins and their Potential Cross-reactivity with Emerging Variants and other Human Coronaviruses
- PMID: 36336501
- PMCID: PMC9633039
- DOI: 10.1016/j.arcmed.2022.10.007
Bioinformatic Analysis of B- and T-cell Epitopes from SARS-CoV-2 Structural Proteins and their Potential Cross-reactivity with Emerging Variants and other Human Coronaviruses
Abstract
Background: The mutations in SARS-CoV-2 variants of concern (VOC) facilitate the virus' escape from the neutralizing antibodies induced by vaccines. However, the protection from hospitalization and death is not significantly diminished. Both vaccine boosters and infection improve immune responses and provide protection, suggesting that conserved and/or cross-reactive epitopes could be involved. While several important T- and B-cell epitopes have been identified, mainly in the S protein, the M and N proteins and their potential cross-reactive epitopes with other coronaviruses remain largely unexplored.
Aims: To identify and map new potential B- and T-cell epitopes within the SARS-CoV-2 S, M and N proteins, as well as cross-reactive epitopes with human coronaviruses.
Methods: Different bioinformatics tools were used to: i) Identify new and compile previously-reported B-and T-cell epitopes from SARS-CoV-2 S, M and N proteins; ii) Determine the mutations in S protein from VOC that affect B- and T-cell epitopes, and; iii) Identify cross-reactive epitopes with coronaviruses relevant to human health.
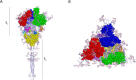
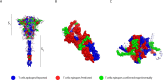
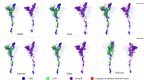
Results: New, potential B- and T-cell epitopes from S, M and N proteins as well as cross-reactive epitopes with other coronaviruses were found and mapped within the proteins' structures.
Conclusion: Numerous potential B- and T-cell epitopes were found in S, M and N proteins, some of which are conserved between coronaviruses. VOCs present mutations within important epitopes in the S protein; however, a significant number of other epitopes remain unchanged. The epitopes identified here may contribute to augmenting the protective response to SARS-CoV-2 and its variants induced by infection and/or vaccination, and may also be used for the rational design of novel broad-spectrum coronavirus vaccines.
Keywords: B- and T-cell epitopes; COVID-19 vaccines.Introduction; Cross-reactivity; Membrane protein; Nucleocapsid protein; SARS-CoV-2; Spike protein.
Copyright © 2022. Published by Elsevier Inc.
Conflict of interest statement
Conflicts of Interest All authors declare no conflicts of interest.
Figures




References
-
- ArcGIS Dashboards 2020. Available from: https://www.arcgis.com/apps/dashboards/bda7594740fd40299423467b48e9ecf6. (Accessed November 22, 2021).
-
- Farmacovigilancia de vacunas para COVID-19 - Catálogo. Farmacovigilancia de vacunas para COVID-19 2022. Available from: https://covid-19pharmacovigilance.paho.org/(Accessed August 10, 2022).
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

