A serralysin-like protein of Candidatus Liberibacter asiaticus modulates components of the bacterial extracellular matrix
- PMID: 36338045
- PMCID: PMC9627510
- DOI: 10.3389/fmicb.2022.1006962
A serralysin-like protein of Candidatus Liberibacter asiaticus modulates components of the bacterial extracellular matrix
Abstract
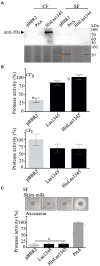
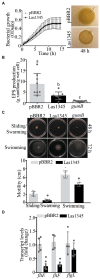
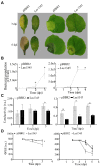
Huanglongbing (HLB), the current major threat for Citrus species, is caused by intracellular alphaproteobacteria of the genus Candidatus Liberibacter (CaL), with CaL asiaticus (CLas) being the most prevalent species. This bacterium inhabits phloem cells and is transmitted by the psyllid Diaphorina citri. A gene encoding a putative serralysin-like metalloprotease (CLIBASIA_01345) was identified in the CLas genome. The expression levels of this gene were found to be higher in citrus leaves than in psyllids, suggesting a function for this protease in adaptation to the plant environment. Here, we study the putative role of CLas-serralysin (Las1345) as virulence factor. We first assayed whether Las1345 could be secreted by two different surrogate bacteria, Rhizobium leguminosarum bv. viciae A34 (A34) and Serratia marcescens. The protein was detected only in the cellular fraction of A34 and S. marcescens expressing Las1345, and increased protease activity of those bacteria by 2.55 and 4.25-fold, respectively. In contrast, Las1345 expressed in Nicotiana benthamiana leaves did not show protease activity nor alterations in the cell membrane, suggesting that Las1345 do not function as a protease in the plant cell. Las1345 expression negatively regulated cell motility, exopolysaccharide production, and biofilm formation in Xanthomonas campestris pv. campestris (Xcc). This bacterial phenotype was correlated with reduced growth and survival on leaf surfaces as well as reduced disease symptoms in N. benthamiana and Arabidopsis. These results support a model where Las1345 could modify extracellular components to adapt bacterial shape and appendages to the phloem environment, thus contributing to virulence.
Keywords: Huanglongbing; biofilm; protease; surrogate bacteria; virulence factor.
Copyright © 2022 Garcia, Molina, Padgett-Pagliai, Torres, Bruna, Garciía Véscovi, González, Gadea and Marano.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
A "Candidatus Liberibacter asiaticus"-secreted polypeptide suppresses plant immune responses in Nicotiana benthamiana and Citrus sinensis.Front Plant Sci. 2022 Oct 24;13:997825. doi: 10.3389/fpls.2022.997825. eCollection 2022. Front Plant Sci. 2022. PMID: 36352861 Free PMC article.
-
Progress and Obstacles in Culturing 'Candidatus Liberibacter asiaticus', the Bacterium Associated with Huanglongbing.Phytopathology. 2019 Jul;109(7):1092-1101. doi: 10.1094/PHYTO-02-19-0051-RVW. Epub 2019 Jun 3. Phytopathology. 2019. PMID: 30998129 Review.
-
'Candidatus Liberibacter americanus', associated with citrus huanglongbing (greening disease) in São Paulo State, Brazil.Int J Syst Evol Microbiol. 2005 Sep;55(Pt 5):1857-1862. doi: 10.1099/ijs.0.63677-0. Int J Syst Evol Microbiol. 2005. PMID: 16166678
-
Distribution and Variation of Bacterial Endosymbiont and "Candidatus Liberibacter asiaticus" Titer in the Huanglongbing Insect Vector, Diaphorina citri Kuwayama.Microb Ecol. 2019 Jul;78(1):206-222. doi: 10.1007/s00248-018-1290-1. Epub 2018 Nov 24. Microb Ecol. 2019. PMID: 30474731
-
Temporal and spatial detection of Candidatus Liberibacter asiaticus putative effector transcripts during interaction with Huanglongbing-susceptible, -tolerant, and -resistant citrus hosts.BMC Plant Biol. 2019 Apr 2;19(1):122. doi: 10.1186/s12870-019-1703-4. BMC Plant Biol. 2019. PMID: 30940073 Free PMC article.
Cited by
-
Recombinant serralysin metalloproteases D enhances the intracellular replication of infectious bovine rhinotracheitis virus.Front Microbiol. 2025 May 9;16:1567288. doi: 10.3389/fmicb.2025.1567288. eCollection 2025. Front Microbiol. 2025. PMID: 40415927 Free PMC article.
References
-
- Alves M. N., Cifuentes-Arenas J. C., Raiol-Junior L. L., Ferro J. A., Peña L. (2021). Early population dynamics of 'Candidatus Liberibacter asiaticus' in susceptible and resistant genotypes after inoculation with infected Diaphorina citri feeding on young shoots. Front. Microbiol. 12:683923. doi: 10.3389/fmicb.2021.683923, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

