Reconstructing neural circuits using multiresolution correlated light and electron microscopy
- PMID: 36338333
- PMCID: PMC9635852
- DOI: 10.3389/fncir.2022.753496
Reconstructing neural circuits using multiresolution correlated light and electron microscopy
Abstract
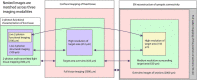
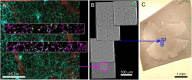
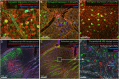
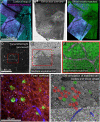
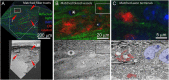
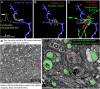
Correlated light and electron microscopy (CLEM) can be used to combine functional and molecular characterizations of neurons with detailed anatomical maps of their synaptic organization. Here we describe a multiresolution approach to CLEM (mrCLEM) that efficiently targets electron microscopy (EM) imaging to optically characterized cells while maintaining optimal tissue preparation for high-throughput EM reconstruction. This approach hinges on the ease with which arrays of sections collected on a solid substrate can be repeatedly imaged at different scales using scanning electron microscopy. We match this multiresolution EM imaging with multiresolution confocal mapping of the aldehyde-fixed tissue. Features visible in lower resolution EM correspond well to features visible in densely labeled optical maps of fixed tissue. Iterative feature matching, starting with gross anatomical correspondences and ending with subcellular structure, can then be used to target high-resolution EM image acquisition and annotation to cells of interest. To demonstrate this technique and range of images used to link live optical imaging to EM reconstructions, we provide a walkthrough of a mouse retinal light to EM experiment as well as some examples from mouse brain slices.
Keywords: confocal 3D microscopy; connectomics; correlated light and electron microscopy (CLEM); electron microscopy; neural circuit; synapse; tissue mapping.
Copyright © 2022 Friedrichsen, Ramakrishna, Hsiang, Valkova, Kerschensteiner and Morgan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

