Multi-omic data integration for the study of production, carcass, and meat quality traits in Nellore cattle
- PMID: 36338989
- PMCID: PMC9634488
- DOI: 10.3389/fgene.2022.948240
Multi-omic data integration for the study of production, carcass, and meat quality traits in Nellore cattle
Abstract
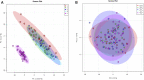
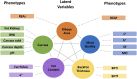
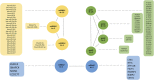
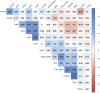
Data integration using hierarchical analysis based on the central dogma or common pathway enrichment analysis may not reveal non-obvious relationships among omic data. Here, we applied factor analysis (FA) and Bayesian network (BN) modeling to integrate different omic data and complex traits by latent variables (production, carcass, and meat quality traits). A total of 14 latent variables were identified: five for phenotype, three for miRNA, four for protein, and two for mRNA data. Pearson correlation coefficients showed negative correlations between latent variables miRNA 1 (mirna1) and miRNA 2 (mirna2) (-0.47), ribeye area (REA) and protein 4 (prot4) (-0.33), REA and protein 2 (prot2) (-0.3), carcass and prot4 (-0.31), carcass and prot2 (-0.28), and backfat thickness (BFT) and miRNA 3 (mirna3) (-0.25). Positive correlations were observed among the four protein factors (0.45-0.83): between meat quality and fat content (0.71), fat content and carcass (0.74), fat content and REA (0.76), and REA and carcass (0.99). BN presented arcs from the carcass, meat quality, prot2, and prot4 latent variables to REA; from meat quality, REA, mirna2, and gene expression mRNA1 to fat content; from protein 1 (prot1) and mirna2 to protein 5 (prot5); and from prot5 and carcass to prot2. The relations of protein latent variables suggest new hypotheses about the impact of these proteins on REA. The network also showed relationships among miRNAs and nebulin proteins. REA seems to be the central node in the network, influencing carcass, prot2, prot4, mRNA1, and meat quality, suggesting that REA is a good indicator of meat quality. The connection among miRNA latent variables, BFT, and fat content relates to the influence of miRNAs on lipid metabolism. The relationship between mirna1 and prot5 composed of isoforms of nebulin needs further investigation. The FA identified latent variables, decreasing the dimensionality and complexity of the data. The BN was capable of generating interrelationships among latent variables from different types of data, allowing the integration of omics and complex traits and identifying conditional independencies. Our framework based on FA and BN is capable of generating new hypotheses for molecular research, by integrating different types of data and exploring non-obvious relationships.
Keywords: Bayesian network; factor analysis; latent variables; meat quality; omics data.
Copyright © 2022 Novais, Yu, Cesar, Momen, Poleti, Petry, Mourão, Regitano, Morota and Coutinho.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Aass L. (1996). Variation in carcass and meat quality traits and their relations to growth in dual purpose cattle. Livest. Prod. Sci. 46, 1–12. 10.1016/0301-6226(96)00005-X - DOI
-
- Adzitey F. (2011). Effect of pre-slaughter animal handling on carcass and meat quality. Int. Food Res. J. 18.
-
- Anderson T. J., Parrish F. C. (1989). Postmortem degradation of titin and nebulin of beef steaks varying in tenderness. J. Food Sci. 54, 748–749. 10.1111/j.1365-2621.1989.tb04695.x - DOI
-
- Andrews S. (2010). FastQC: A quality control tool for high throughput sequence data.
LinkOut - more resources
Full Text Sources
Research Materials

