NGS in the clinical microbiology settings
- PMID: 36339334
- PMCID: PMC9627026
- DOI: 10.3389/fcimb.2022.955481
NGS in the clinical microbiology settings
Abstract
We hypothesized that targeted NGS sequencing might have an advantage over Sanger sequencing, especially in polymicrobial infections. The study included 55 specimens from 51 patients. We compared targeted NGS to Sanger sequencing in clinical samples submitted for Sanger sequencing. The overall concordance rate was 58% (32/55) for NGS vs. Sanger. NGS identified 9 polymicrobial and 2 monomicrobial infections among 19 Sanger-negative samples and 8 polymicrobial infections in 11 samples where a 16S gene was identified by gel electrophoresis, but could not be mapped to an identified pathogen by Sanger. We estimated that NGS could have contributed to patient management in 6/18 evaluated patients and thus has an advantage over Sanger sequencing in certain polymicrobial infections.
Keywords: 16s; NGS; clinical microbiology; next generation sequencing; polymicrobial; polymicrobial infections.
Copyright © 2022 Pitashny, Kadry, Shalaginov, Gazit, Zohar, Szwarcwort, Stabholz and Paul.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
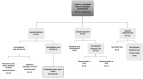
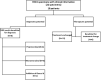
References
-
- Apprill A., Mcnally S., Parsons R., Weber L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat Microb. Ecol. 75 (2), 129–137.
-
- Culbreath K., Melanson S., Gale J., Baker J., Li F., Saebo O., et al. . (2019). Validation and retrospective clinical evaluation of a quantitative 16S rRNA gene metagenomic sequencing assay for bacterial pathogen detection in body fluids. J. Mol. Diagn. 21 (5), 913–923. - PubMed
-
- Khoury N., Amit S., Geffen Y., Adler A. (2019). Clinical utility of pan-microbial PCR assays in the routine diagnosis of infectious diseases. Diagn. Microbiol. Infect. Dis. 93 (3), 232–237. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

