m6A-Related Genes Contribute to Poor Prognosis of Hepatocellular Carcinoma
- PMID: 36339682
- PMCID: PMC9629938
- DOI: 10.1155/2022/2427987
m6A-Related Genes Contribute to Poor Prognosis of Hepatocellular Carcinoma
Abstract
Background: Hepatocellular carcinoma (HCC) is one of the most common and lethal digestive system cancers worldwide. N6-methyladenosine (m6A) modification plays an essential role in diverse critical biological processes and may participate in the development and progression of HCC.
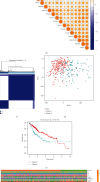
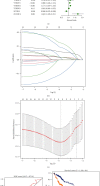
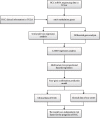
Methods: We downloaded transcriptome data and clinical data from TCGA as the training set. COX and LASSO screened prognostic m6A genes. ROC and Kaplan-Meier curve analysis evaluated the effectiveness of the model. ICGC and our center data were used as verification sets.
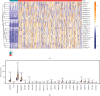
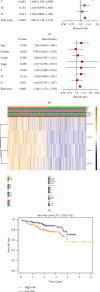
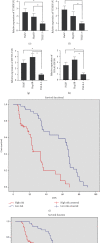
Results: We include the "writer (METTL3, METTL14, WTAP, KIAA1429, RBM15, ZC3H13)," the "reader (YTHDC1, YTHDC2, YTHDF1, YTHDF2, HNRNPC)," and the "eraser (FTO, ALKBH5)" in the study. We obtained YTHDF2, YTHDF1, METTL3, and KIAA1429 through differential analysis, survival analysis, and LASSO regression analysis. The prediction model was established based on the expression of these 4 molecules. HCC patients were divided into "high-risk" and "low-risk" groups to compare survival differences. The model suggested a poor prognosis in the validation sets.
Conclusion: The four-m6A-related-gene combination model was an independent prognostic factor of HCC and could improve the prediction of the prognosis of HCC.
Copyright © 2022 Yan Zou et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

