Identifying tumor immunity-associated molecular features in liver hepatocellular carcinoma by multi-omics analysis
- PMID: 36339710
- PMCID: PMC9632276
- DOI: 10.3389/fmolb.2022.960457
Identifying tumor immunity-associated molecular features in liver hepatocellular carcinoma by multi-omics analysis
Abstract
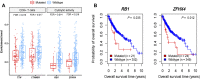
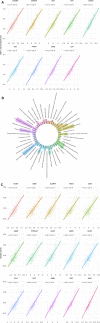
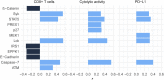
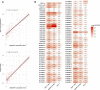
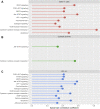
Background: Although current immunotherapies have achieved some successes for hepatocellular carcinoma (HCC) patients, their benefits are limited for most HCC patients. Therefore, the identification of biomarkers for promoting immunotherapeutic responses in HCC is urgently needed. Methods: Using the TCGA HCC cohort, we investigated correlations of various molecular features with antitumor immune signatures (CD8+ T cell infiltration and cytolytic activity) and an immunosuppressive signature (PD-L1 expression) in HCC. These molecular features included mRNAs, microRNAs (miRNAs), long non-coding RNAs (lncRNAs), proteins, and pathways. Results: We found that the mutations of several oncogenes and tumor suppressor genes significantly correlated with reduced antitumor immune signatures, including TTN, CTNNB1, RB1, ZFHX4, and TP53. It indicates that these genes' mutations may inhibit antitumor immune responses in HCC. Four proteins (Syk, Lck, STAT5, and Caspase-7) had significant positive expression correlations with CD8+ T cell enrichment, cytolytic activity, and PD-L1 expression in HCC. It suggests that these proteins' expression could be useful biomarkers for the response to immune checkpoint inhibitors Similiarly, we identified other types of biomarkers potentially useful for predicting the response to ICIs, including miRNAs (hsa-miR-511-5p, 150-3p, 342-3p, 181a-3p, 625-5p, 4772-3p, 155-3p, 142-5p, 142-3p, 155-5p, 625-3p, 1976, 7702), many lncRNAs, and pathways (apoptosis, cytokine-cytokine receptor interaction, Jak-STAT signaling, MAPK signaling, PI3K-AKT signaling, HIF-1 signaling, ECM receptor interaction, focal adhesion, and estrogen signaling). Further, tumor mutation burden showed no significant correlation with antitumor immunity, while tumor aneuploidy levels showed a significant negative correlation with antitumor immunity. Conclusion: The molecular features significantly associated with HCC immunity could be predictive biomarkers for immunotherapeutic responses in HCC patients. They could also be potential intervention targets for boosting antitumor immunity and immunotherapeutic responses in HCC.
Keywords: antitumor immunity; biomarker; cancer immunotherapy; hepatocellular carcinoma; multi-omics analysis.
Copyright © 2022 Shen, He, Qian and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300. 10.1111/j.2517-6161.1995.tb02031.x - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

