Differential Expression and Bioinformatics Analysis of Plasma-Derived Exosomal circRNA in Type 1 Diabetes Mellitus
- PMID: 36339941
- PMCID: PMC9634467
- DOI: 10.1155/2022/3625052
Differential Expression and Bioinformatics Analysis of Plasma-Derived Exosomal circRNA in Type 1 Diabetes Mellitus
Abstract
Backgrounds: Both exosome and circular RNA (circRNA) have been reported to participate in the pathogenesis of type 1 diabetes mellitus (T1DM). However, the exact role of exosomal circRNA in T1DM is largely unknown. Here, we identified the exosomal circRNA expression profiles in the plasma of T1DM patients and explored their potential function using bioinformatics analysis. Material and Methods. Exosomes were extracted by the size exclusion chromatography method from plasma of 10 T1DM patients and 10 age- and sex- matched control subjects. Illumina Novaseq6000 platform was used to detect the exosomal circRNA expression profiles. Multiple bioinformatics analysis was applied to investigate the potential biological functions of exosomal circRNAs.
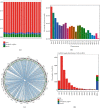
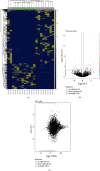
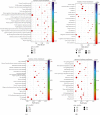
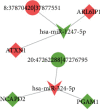
Results: A total of 784 differentially expressed exosomal circRNAs have been identified in T1DM patients, of which 528 were upregulated and 256 were downregulated. Gene Ontology analysis enriched terms such as protein ubiquitination involved in ubiquitin-dependent protein catabolic protein (GO:0042787), membrane (GO:0016020), and GTPase activator activity (GO:0005096). The most enriched pathway in Kyoto Encyclopedia of Genes and Genomes was ubiquitin-mediated proteolysis (ko04120). The miRNA-targeting prediction method was used to identify the miRNAs that bind to circRNAs, and circRNA-miRNA-mRNA pathways were constructed, indicating that interactions between circRNA, miRNA, and gene might be involved in the disease progression.
Conclusions: The present study identified the exosomal circRNA expression profiles in T1DM for the first time. Our results threw novel insights into the molecular mechanisms of T1DM.
Copyright © 2022 Haipeng Pang et al.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Analysis of expression profiles and bioinformatics suggests that plasma exosomal circular RNAs may be involved in ischemic stroke in the Chinese Han population.Metab Brain Dis. 2022 Mar;37(3):665-676. doi: 10.1007/s11011-021-00894-2. Epub 2022 Jan 24. Metab Brain Dis. 2022. PMID: 35067794
-
Plasma-derived exosomal mRNA profiles associated with type 1 diabetes mellitus.Front Immunol. 2022 Sep 13;13:995610. doi: 10.3389/fimmu.2022.995610. eCollection 2022. Front Immunol. 2022. PMID: 36177022 Free PMC article.
-
Characterization of lncRNA Profiles of Plasma-Derived Exosomes From Type 1 Diabetes Mellitus.Front Endocrinol (Lausanne). 2022 May 12;13:822221. doi: 10.3389/fendo.2022.822221. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 35634499 Free PMC article.
-
Presence and prospects of exosomal circRNAs in cancer (Review).Int J Oncol. 2023 Apr;62(4):47. doi: 10.3892/ijo.2023.5495. Epub 2023 Mar 3. Int J Oncol. 2023. PMID: 36866755 Review.
-
Exosomal circRNA: emerging insights into cancer progression and clinical application potential.J Hematol Oncol. 2023 Jun 26;16(1):67. doi: 10.1186/s13045-023-01452-2. J Hematol Oncol. 2023. PMID: 37365670 Free PMC article. Review.
Cited by
-
Exosomes; a Potential Source of Biomarkers, Therapy, and Cure for Type-1 Diabetes.Int J Mol Sci. 2023 Oct 28;24(21):15713. doi: 10.3390/ijms242115713. Int J Mol Sci. 2023. PMID: 37958696 Free PMC article. Review.
-
CircRNAs and RNA-Binding Proteins Involved in the Pathogenesis of Cancers or Central Nervous System Disorders.Noncoding RNA. 2023 Mar 31;9(2):23. doi: 10.3390/ncrna9020023. Noncoding RNA. 2023. PMID: 37104005 Free PMC article. Review.
-
Exosomes in Autoimmune Diseases: A Review of Mechanisms and Diagnostic Applications.Clin Rev Allergy Immunol. 2025 Jan 17;68(1):5. doi: 10.1007/s12016-024-09013-2. Clin Rev Allergy Immunol. 2025. PMID: 39820756 Review.
-
The Emerging Role of Natural Products in Cancer Treatment.Arch Toxicol. 2024 Aug;98(8):2353-2391. doi: 10.1007/s00204-024-03786-3. Epub 2024 May 25. Arch Toxicol. 2024. PMID: 38795134 Review.
-
Importance of Studying Non-Coding RNA in Children and Adolescents with Type 1 Diabetes.Biomedicines. 2024 Sep 2;12(9):1988. doi: 10.3390/biomedicines12091988. Biomedicines. 2024. PMID: 39335501 Free PMC article. Review.
References
-
- Patterson C. C., Karuranga S., Salpea P., et al. Worldwide estimates of incidence, prevalence and mortality of type 1 diabetes in children and adolescents: results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Research and Clinical Practice . (9th edition) 2019;157 doi: 10.1016/j.diabres.2019.107842. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

