Left-right asymmetric expression of the Nodal-Lefty-Pitx2 module in developing turtle forebrain
- PMID: 36340044
- PMCID: PMC9634164
- DOI: 10.3389/fcell.2022.929808
Left-right asymmetric expression of the Nodal-Lefty-Pitx2 module in developing turtle forebrain
Abstract
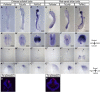
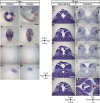
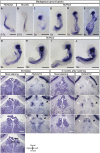
The epithalamus of zebrafish shows morphological and molecular left-right (L-R) asymmetry, but such asymmetry is not apparent in tetrapods. To provide further insight into the evolutionary diversity of brain L-R asymmetry, we have now examined the developing brains of reptile embryos for expression of Nodal, Lefty, and Pitx2. Two turtle species, the Chinese softshell turtle and the red-eared slider turtle, showed left-sided expression of these three genes in the developing forebrain, with this expression occurring after Nodal expression at the lateral plate and the L-R organizer has disappeared. Nodal activity, as revealed by the detection of phosphorylated Smad2/3, was also apparent in the neural epithelium on the left side in both turtle species. In the Chinese softshell turtle, the habenula did not show apparent asymmetry in size and the parapineal organ was absent, but the expression of Kctd12 in the habenula showed a small yet reproducible asymmetry. In contrast to the turtles, L-R asymmetric expression of Nodal, Lefty, Pitx2, or Kctd12 was not detected in the developing brain of the Madagascar ground gecko. The transcriptional enhancer (ASE) responsible for the asymmetric expression of Nodal, Lefty, and Pitx2 was conserved among reptiles, including the Chinese softshell turtle and Madagascar ground gecko. Our findings suggest that Nodal, Lefty, and Pitx2 have the potential to be asymmetrically expressed in the developing brain of vertebrates, but that their expression varies even among reptiles.
Keywords: Nodal; brain; left-right asymmetry; reptile; turtle.
Copyright © 2022 Kajikawa, Miki, Takeda, Kiyonari and Hamada.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Nodal paralogues underlie distinct mechanisms for visceral left-right asymmetry in reptiles and mammals.Nat Ecol Evol. 2020 Feb;4(2):261-269. doi: 10.1038/s41559-019-1072-2. Epub 2020 Jan 6. Nat Ecol Evol. 2020. PMID: 31907383
-
Determination of left/right asymmetric expression of nodal by a left side-specific enhancer with sequence similarity to a lefty-2 enhancer.Genes Dev. 1999 Jun 15;13(12):1589-600. doi: 10.1101/gad.13.12.1589. Genes Dev. 1999. PMID: 10385627 Free PMC article.
-
Smad5 is essential for left-right asymmetry in mice.Dev Biol. 2000 Mar 1;219(1):71-8. doi: 10.1006/dbio.1999.9594. Dev Biol. 2000. PMID: 10677256
-
Asymmetry in the epithalamus of vertebrates.J Anat. 2001 Jul-Aug;199(Pt 1-2):63-84. doi: 10.1046/j.1469-7580.2001.19910063.x. J Anat. 2001. PMID: 11523830 Free PMC article. Review.
-
TGFβ signaling in establishing left-right asymmetry.Semin Cell Dev Biol. 2014 Aug;32:80-4. doi: 10.1016/j.semcdb.2014.03.029. Epub 2014 Apr 2. Semin Cell Dev Biol. 2014. PMID: 24704359 Review.
Cited by
-
Cdon is essential for organ left-right patterning by regulating dorsal forerunner cells clustering and Kupffer's vesicle morphogenesis.Front Cell Dev Biol. 2024 Aug 22;12:1429782. doi: 10.3389/fcell.2024.1429782. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39239564 Free PMC article.
References
LinkOut - more resources
Full Text Sources

