Biosensor Cell-Fit-HD4D for correlation of single-cell fate and microscale energy deposition in complex ion beams
- PMID: 36340882
- PMCID: PMC9627659
- DOI: 10.1016/j.xpro.2022.101798
Biosensor Cell-Fit-HD4D for correlation of single-cell fate and microscale energy deposition in complex ion beams
Abstract
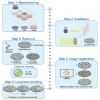
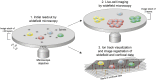
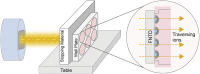
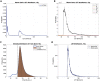
We present a protocol for the biosensor Cell-Fit-HD4D. It enables long-term monitoring and correlation of single-cell fate with subcellular-deposited energy of ionizing radiation. Cell fate tracking using widefield time-lapse microscopy is uncoupled in time from confocal ion track imaging. Registration of both image acquisition steps allows precise ion track assignment to cells and correlation with cellular readouts. For complete details on the use and execution of this protocol, please refer to Niklas et al. (2022).
Keywords: Biophysics; Biotechnology and bioengineering; Cancer; Microscopy; Molecular biology; Molecular/Chemical probes; Single cell.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Akselrod M., Kouwenberg J. Fluorescent nuclear track detectors - review of past, present and future of the technology. Radiat. Meas. 2018;117:35–51. doi: 10.1016/j.radmeas.2018.07.005. - DOI
-
- Akselrod M.S., Sykora G.J. Fluorescent nuclear track detector technology – a new way to do passive solid state dosimetry. Radiat. Meas. 2011;46:1671–1679. doi: 10.1016/j.radmeas.2011.06.018. - DOI
-
- Dokic I., Mairani A., Niklas M., Zimmermann F., Chaudhri N., Krunic D., Tessonnier T., Ferrari A., Parodi K., Jäkel O., et al. Next generation multi-scale biophysical characterization of high precision cancer particle radiotherapy using clinical proton, helium-carbon- and oxygen ion beams. Oncotarget. 2016;7:56676–56689. doi: 10.18632/oncotarget.10996. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

