Identification and validation of a tyrosine metabolism-related prognostic prediction model and characterization of the tumor microenvironment infiltration in hepatocellular carcinoma
- PMID: 36341373
- PMCID: PMC9633179
- DOI: 10.3389/fimmu.2022.994259
Identification and validation of a tyrosine metabolism-related prognostic prediction model and characterization of the tumor microenvironment infiltration in hepatocellular carcinoma
Abstract
Background: Hepatocellular carcinoma (HCC) is an aggressive and heterogeneous disease characterized by high morbidity and mortality. The liver is the vital organ that participates in tyrosine catabolism, and abnormal tyrosine metabolism could cause various diseases, including HCC. Besides, the tumor immune microenvironment is involved in carcinogenesis and can influence the patients' clinical outcomes. However, the potential role of tyrosine metabolism pattern and immune molecular signature is poorly understood in HCC.
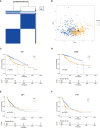
Methods: Gene expression, somatic mutations, copy number variation data, and clinicopathological information of HCC were downloaded from The Cancer Genome Atlas (TCGA) database. GSE14520 from the Gene Expression Omnibus (GEO) databases was used as a validation dataset. We performed unsupervised consensus clustering of tyrosine metabolism-related genes (TRGs) and classified patients into distinct molecular subtypes. We used ESTIMATE algorithms to evaluate the immune infiltration. We then applied LASSO Cox regression to establish the TRGs risk model and validated its predictive performance.
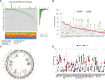
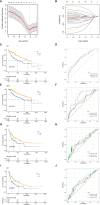
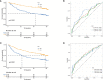
Results: In this study, we first described the alterations of 42 TRGs in HCC cohorts and characterized the clinicopathological characteristics and tumor microenvironmental landscape of the two distinct subtypes. We then established a tyrosine metabolism-related scoring system and identified five TRGs, which were highly correlated with prognosis and representative of this gene set, namely METTL6, GSTZ1, ADH4, ADH1A, and LCMT1. Patients in the high-risk group had an inferior prognosis. Univariate and multivariate Cox proportional hazards regression analysis also showed that the tyrosine metabolism-related signature was an independent prognostic indicator. Besides, receiver operating characteristic curve (ROC) analysis demonstrated the predictive accuracy of the TRGs signature that could reliably predict 1-, 3-, and 5-year survival in both TCGA and GEO cohorts. We also got consistent results by performing clone formation and invasion analysis, and immunohistochemical (IHC) assays. Moreover, we also discovered that the TRGs signature was significantly associated with the different immune landscapes and therapeutic drug sensitivity.
Conclusion: Our comprehensive analysis revealed the potential molecular signature and clinical utilities of TRGs in HCC. The model based on five TRGs can accurately predict the survival outcomes of HCC, improving our knowledge of TRGs in HCC and paving a new path for guiding risk stratification and treatment strategy development for HCC patients.
Keywords: hepatocellular carcinoma; immunotherapy; prognosis model; tumor microenvironment; tyrosine metabolism.
Copyright © 2022 Zhou, Li, Long, Tao, Zhou and Tang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

