Humoral immunity and transcriptome differences of COVID-19 inactivated vacciane and protein subunit vaccine as third booster dose in human
- PMID: 36341453
- PMCID: PMC9634958
- DOI: 10.3389/fimmu.2022.1027180
Humoral immunity and transcriptome differences of COVID-19 inactivated vacciane and protein subunit vaccine as third booster dose in human
Abstract
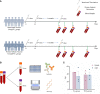
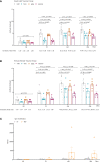
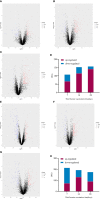
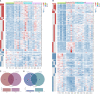
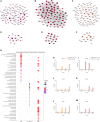
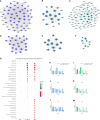
Under the background of the severe human health and world economic burden caused by COVID-19, the attenuation of vaccine protection efficacy, and the prevalence and immune escape of emerging variants of concern (VOCs), the third dose of booster immunization has been put on the agenda. Systems biology approaches can help us gain new perspectives on the characterization of immune responses and the identification of factors underlying vaccine-induced immune efficacy. We analyzed the antibody signature and transcriptional responses of participants vaccinated with COVID-19 inactivated vaccine and protein subunit vaccine as a third booster dose. The results from the antibody indicated that the third booster dose was effective, and that heterologous vaccination with the protein subunit vaccine as a booster dose induced stronger humoral immune responses than the homologous vaccination with inactivated vaccine, and might be more effective against VOCs. In transcriptomic analysis, protein subunit vaccine induced more differentially expressed genes that were significantly associated with many important innate immune pathways. Both the homologous and heterologous boosters could increase the effectiveness against COVID-19, and compared with the inactivated vaccine, the protein subunit vaccine, mediated a stronger humoral immune response and had a more significant correlation with the innate immune function module, which provided certain data support for the third booster immunization strategy.
Keywords: COVID-19; humoral immunity; third booster vaccine; transcriptome analysis; variants of concern.
Copyright © 2022 Zhang, Yao, Guo, Han, Zhang, Zhang, Jiang, Wang, Fang, Wang, Pang, Liu, Kou and Jiang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Immune responses and transcription landscape of adults with the third dose of homologous and heterologous booster vaccines of COVID-19.Front Immunol. 2024 Sep 12;15:1461419. doi: 10.3389/fimmu.2024.1461419. eCollection 2024. Front Immunol. 2024. PMID: 39328415 Free PMC article.
-
Persistence of immunity and impact of third dose of inactivated COVID-19 vaccine against emerging variants.Sci Rep. 2022 Jul 14;12(1):12038. doi: 10.1038/s41598-022-16097-3. Sci Rep. 2022. PMID: 35835822 Free PMC article. Clinical Trial.
-
[Humoral Immune Response in Inactivated SARS-CoV-2 Vaccine: When Should a Booster Dose be Administered?].Mikrobiyol Bul. 2022 Jul;56(3):566-573. doi: 10.5578/mb.20229715. Mikrobiyol Bul. 2022. PMID: 35960246 Turkish.
-
Characterization of SARS-CoV-2-Specific Humoral and Cellular Immune Responses Induced by Inactivated COVID-19 Vaccines in a Real-World Setting.Front Immunol. 2021 Dec 22;12:802858. doi: 10.3389/fimmu.2021.802858. eCollection 2021. Front Immunol. 2021. PMID: 35003131 Free PMC article.
-
Booster vaccination strategy: Necessity, immunization objectives, immunization strategy, and safety.J Med Virol. 2022 Jun;94(6):2369-2375. doi: 10.1002/jmv.27590. Epub 2022 Jan 19. J Med Virol. 2022. PMID: 35028946 Review.
Cited by
-
Immune responses and transcription landscape of adults with the third dose of homologous and heterologous booster vaccines of COVID-19.Front Immunol. 2024 Sep 12;15:1461419. doi: 10.3389/fimmu.2024.1461419. eCollection 2024. Front Immunol. 2024. PMID: 39328415 Free PMC article.
-
Antibody and transcription landscape in peripheral blood mononuclear cells of elderly adults over 70 years of age with third dose of COVID-19 BBIBP-CorV and ZF2001 booster vaccine.Immun Ageing. 2024 Jan 27;21(1):11. doi: 10.1186/s12979-023-00408-x. Immun Ageing. 2024. PMID: 38280989 Free PMC article.
References
-
- Brisotto G, Muraro E, Montico M, Corso C, Evangelista C, Casarotto M, et al. . IgG antibodies against SARS-CoV-2 decay but persist 4 months after vaccination in a cohort of healthcare workers. Clinica chimica acta; Int J Clin Chem (2021) 523:476–82. doi: 10.1016/j.cca.2021.10.035 - DOI - PMC - PubMed
-
- Vicenti I, Basso M, Gatti F, Scaggiante R, Boccuto A, Zago D, et al. . Faster decay of neutralizing antibodies in never infected than previously infected healthcare workers three months after the second BNT162b2 mRNA COVID-19 vaccine dose. Int J Infect Dis IJID Off Publ Int Soc Infect Dis (2021) 112:40–4. doi: 10.1016/j.ijid.2021.08.052 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

