Microbial dysbiosis in the gut drives systemic autoimmune diseases
- PMID: 36341463
- PMCID: PMC9632986
- DOI: 10.3389/fimmu.2022.906258
Microbial dysbiosis in the gut drives systemic autoimmune diseases
Abstract
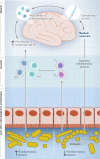
Trillions of microbes survive and thrive inside the human body. These tiny creatures are crucial to the development and maturation of our immune system and to maintain gut immune homeostasis. Microbial dysbiosis is the main driver of local inflammatory and autoimmune diseases such as colitis and inflammatory bowel diseases. Dysbiosis in the gut can also drive systemic autoimmune diseases such as type 1 diabetes, rheumatic arthritis, and multiple sclerosis. Gut microbes directly interact with the immune system by multiple mechanisms including modulation of the host microRNAs affecting gene expression at the post-transcriptional level or production of microbial metabolites that interact with cellular receptors such as TLRs and GPCRs. This interaction modulates crucial immune functions such as differentiation of lymphocytes, production of interleukins, or controlling the leakage of inflammatory molecules from the gut to the systemic circulation. In this review, we compile and analyze data to gain insights into the underpinning mechanisms mediating systemic autoimmune diseases. Understanding how gut microbes can trigger or protect from systemic autoimmune diseases is crucial to (1) tackle these diseases through diet or lifestyle modification, (2) develop new microbiome-based therapeutics such as prebiotics or probiotics, (3) identify diagnostic biomarkers to predict disease risk, and (4) observe and intervene with microbial population change with the flare-up of autoimmune responses. Considering the microbiome signature as a crucial player in systemic autoimmune diseases might hold a promise to turn these untreatable diseases into manageable or preventable ones.
Keywords: MS; SLE; T1D; arthrities; autoimmune diseases; dysbiosis; microbiome.
Copyright © 2022 Mousa, Chehadeh and Husband.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

