Plasmids for Controlled and Tunable High-Level Expression in E. coli
- PMID: 36342148
- PMCID: PMC9680613
- DOI: 10.1128/aem.00939-22
Plasmids for Controlled and Tunable High-Level Expression in E. coli
Abstract
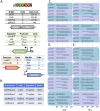
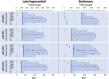
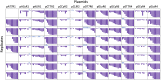
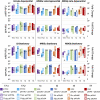
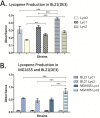
Controlled gene expression is crucial for engineering bacteria for basic and applied research. Inducible systems enable tight regulation of expression, wherein a small-molecule inducer causes the transcription factor to activate or repress transcriptional initiation. The T7 expression system is one of the most widely used inducible systems, particularly for high overexpression of proteins. However, it is well known that the highly active T7 RNA polymerase (RNAP) has several drawbacks, including toxicity to the host and substantial leaky expression in the absence of an inducer. Much work has been done to address these issues; current solutions require special strains or additional plasmids, making the system more complicated and less accessible. Here, we challenge the assumption that the T7 expression system is the best choice for obtaining high protein titers. We hypothesized that expression from strong inducible promoters expressed from high-copy plasmids could compete with expression levels obtained from T7 RNAP but that such promoters would possess improved control of transcription. Employing inducible systems from a toolbox we developed previously, we demonstrate that our plasmids consistently give higher outputs and greater fold changes over basal expression than the T7 system across rich and minimal media. In addition, we show that they outperformed the T7 system when we used an engineered metabolic pathway to produce lycopene. IMPORTANCE Genetic systems for protein overexpression are required tools in microbiological and biochemical research. Ideally, these systems include standardized genetic parts with predictable behavior, enabling the construction of stable expression systems in the host organism. Modularity of a genetic system is advantageous, so that the expression system can be easily moved into a host that best suits the needs of a given experiment. The T7 expression system lacks both predictability and stability and requires special host strains to function. Despite these limitations, it remains one of the most popular systems for protein overproduction. This study directly compared the T7 system to four inducible systems from our broad-host-range plasmid toolbox and demonstrated these alternative expression systems have distinct advantages over the T7. The systems are entirely plasmid-based and not constrained to a specific bacterial host, expanding the options for high-level protein expression across strains.
Keywords: T7 system; inducible expression; plasmid toolbox; protein overexpression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

