Parallel Evolution of Pseudomonas aeruginosa during a Prolonged ICU-Infection Outbreak
- PMID: 36342287
- PMCID: PMC9769503
- DOI: 10.1128/spectrum.02743-22
Parallel Evolution of Pseudomonas aeruginosa during a Prolonged ICU-Infection Outbreak
Abstract
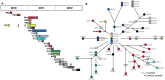
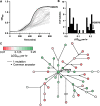
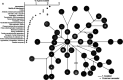
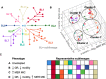
Most knowledge about Pseudomonas aeruginosa pathoadaptation is derived from studies on airway colonization in cystic fibrosis; little is known about adaptation in acute settings. P. aeruginosa frequently affects burned patients and the burn wound niche has distinct properties that likely influence pathoadaptation. This study aimed to genetically and phenotypically characterize P. aeruginosa isolates collected during an outbreak of infection in a burn intensive care unit (ICU). Sequencing reads from 58 isolates of ST1076 P. aeruginosa taken from 23 patients were independently mapped to a complete reference genome for the lineage (H25338); genetic differences were identified and were used to define the population structure. Comparative genomic analysis at single-nucleotide resolution identified pathoadaptive genes that evolved multiple, independent mutations. Three key phenotypic assays (growth performance, motility, carbapenem resistance) were performed to complement the genetic analysis for 47 unique isolates. Population structure for the ST1076 lineage revealed 11 evolutionary sublineages. Fifteen pathoadaptive genes evolved mutations in at least two sublineages. The most prominent functional classes affected were transcription/two-component regulatory systems, and chemotaxis/motility and attachment. The most frequently mutated gene was oprD, which codes for outer membrane porin involved in uptake of carbapenems. Reduced growth performance and motility were found to be adaptive phenotypic traits, as was high level of carbapenem resistance, which correlated with higher carbapenem consumption during the outbreak. Multiple prominent linages evolved each of the three traits in parallel providing evidence that they afford a fitness advantage for P. aeruginosa in the context of human burn infection. IMPORTANCE Pseudomonas aeruginosa is a Gram-negative pathogen causing infections in acutely burned patients. The precise mechanisms required for the establishment of infection in the burn setting, and adaptive traits underpinning prolonged outbreaks are not known. We have assessed genotypic data from 58 independent P. aeruginosa isolates taken from a single lineage that was responsible for an outbreak of infection in a burn ICU that lasted for almost 2.5 years and affected 23 patients. We identified a core set of 15 genes that we predict to control pathoadaptive traits in the burn infection based on the frequency with which independent mutations evolved. We combined the genotypic data with phenotypic data (growth performance, motility, antibiotic resistance) and clinical data (antibiotic consumption) to identify adaptive phenotypes that emerged in parallel. High-level carbapenem resistance evolved rapidly, and frequently, in response to high clinical demand for this antibiotic class during the outbreak.
Keywords: adaptative evolution; antibiotic resistance; carbapenems; oprD; outer membrane porin D.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Jorth P, Durfey S, Rezayat A, Garudathri J, Ratjen A, Staudinger BJ, Radey MC, Genatossio A, McNamara S, Cook DA, Aitken ML, Gibson RL, Yahr TL, Singh PK. 2021. Cystic fibrosis lung function decline after within-host evolution increases virulence of infecting Pseudomonas aeruginosa. Am J Respir Crit Care Med 203:637–640. doi: 10.1164/rccm.202007-2735LE. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

