Identification of novel meQTLs strongly associated with rheumatoid arthritis by large-scale epigenome-wide analysis
- PMID: 36342317
- PMCID: PMC9714356
- DOI: 10.1002/2211-5463.13517
Identification of novel meQTLs strongly associated with rheumatoid arthritis by large-scale epigenome-wide analysis
Abstract
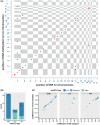
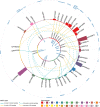
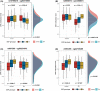
Rheumatoid arthritis (RA) is highly heritable, and previous studies have suggested that genetic variation may affect susceptibility to RA by altering epigenetic modifications (e.g. DNA methylation). Here we examined how genetic variation influences DNA methylation (DNAm) in RA by integrating individual genetic variation and DNAm data. Epigenome-wide meQTL (methylation quantitative trait loci) analysis was performed on 354 RA patients and 335 controls, scanning 30,101,744 relationships between 62 SNPs and 485,512 DNA methylation sites. Two regulatory relationship pairs (FDR < 0.05) showed very strong associations with RA risk. One was rs10796216-cg00475509, and the DNAm decreased by 0.0168 per addition of allele rs10796216-A. The other was rs6546473-cg13358873, for which a 0.0365 reduction of DNAm at cg13358873 was observed for each addition of allele rs6546473-A, and lower DNAm was found to be significantly associated with RA risk (P = 2.0407e-28). Moreover, both pairs of meQTL showed a strong regulatory relationship only in RA samples, so they can be subsequently considered as risk markers for RA. In conclusion, our integrated analysis of genetic and epigenetic variation suggests that genetic variation may affect the risk of RA by regulating DNA methylation levels. Alterations of DNAm at cg00475509 and cg13358873 loci conferred by rs10796216-A and rs6546473-A allele may suggest a potential risk for RA. Our results deepen our understanding of the genetic and epigenetic mechanisms of RA and provide novel associations that can be further investigated in future studies.
Keywords: DNA methylation; SNP; bioinformatics; epigenetics; genetic risk; rheumatoid arthritis.
© 2022 The Authors. FEBS Open Bio published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Epigenome-Wide Association Study of Asthma Exacerbations in Europeans.Allergy. 2025 Apr;80(4):1086-1099. doi: 10.1111/all.16490. Epub 2025 Feb 5. Allergy. 2025. PMID: 39907155
-
meQTL and ncRNA functional analyses of 102 GWAS-SNPs associated with depression implicate HACE1 and SHANK2 genes.Clin Epigenetics. 2020 Jul 2;12(1):99. doi: 10.1186/s13148-020-00884-8. Clin Epigenetics. 2020. PMID: 32616021 Free PMC article.
-
Genetic variants shape rheumatoid arthritis-specific transcriptomic features in CD4+ T cells through differential DNA methylation, explaining a substantial proportion of heritability.Ann Rheum Dis. 2021 Jul;80(7):876-883. doi: 10.1136/annrheumdis-2020-219152. Epub 2021 Jan 12. Ann Rheum Dis. 2021. PMID: 33436383
-
Interplay between genetic and epigenetic mechanisms in rheumatoid arthritis.Epigenomics. 2017 Apr;9(4):493-504. doi: 10.2217/epi-2016-0142. Epub 2017 Mar 21. Epigenomics. 2017. PMID: 28322583 Review.
-
Epigenetics in the pathogenesis of RA.Semin Immunopathol. 2017 Jun;39(4):409-419. doi: 10.1007/s00281-017-0621-5. Epub 2017 Mar 21. Semin Immunopathol. 2017. PMID: 28324153 Review.
References
-
- MacGregor AJ, Snieder H, Rigby AS, Koskenvuo M, Kaprio J, Aho K, et al. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 2000;43:30–7. - PubMed
-
- Dedmon LE. The genetics of rheumatoid arthritis. Rheumatology (Oxford). 2020;59:2661–70. - PubMed
-
- Ding J, Orozco G. Identification of rheumatoid arthritis causal genes using functional genomics. Scand J Immunol. 2019;89:e12753. - PubMed
-
- Nabi G, Akhter N, Wahid M, Bhatia K, Mandal RK, Dar SA, et al. Meta‐analysis reveals PTPN22 1858C/T polymorphism confers susceptibility to rheumatoid arthritis in Caucasian but not in Asian population. Autoimmunity. 2016;49:197–210. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

