The deubiquitylase USP7 is a novel cyclin F-interacting protein and regulates cyclin F protein stability
- PMID: 36342772
- PMCID: PMC9699750
- DOI: 10.18632/aging.204372
The deubiquitylase USP7 is a novel cyclin F-interacting protein and regulates cyclin F protein stability
Abstract
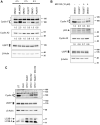
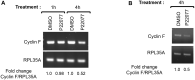
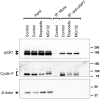
Cyclin F, unlike canonical and transcriptional cyclins, does not bind or activate any cyclin-dependent kinases. Instead, it harbors an F-box motif and primarily functions as the substrate recognition subunit of the Skp1-Cul1-F-box E3 ubiquitin ligase complex, SCFCyclin F. By targeting specific proteins for ubiquitin-mediated proteasomal degradation, cyclin F plays a critical role in the regulation of centrosomal duplication, DNA replication and repair, and maintenance of genomic stability. Cyclin F abundance and activity are tightly regulated throughout the cell cycle. However, the molecular mechanisms regulating cyclin F are scantily understood. Here, we identify the deubiquitylase USP7 as a novel cyclin F-interacting protein. We observe that USP7 stabilizes cyclin F protein and that this function is independent of the deubiquitylase activity of USP7. Additionally, our data suggest that USP7 is also involved in the regulation of cyclin F mRNA. Pharmacological inhibition of the deubiquitylase activity of USP7 resulted in downregulation of cyclin F mRNA.
Keywords: USP7; atypical cyclins; cell cycle; cyclin F; genomic integrity.
Conflict of interest statement
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

