Probabilistic program inference in network-based epidemiological simulations
- PMID: 36342957
- PMCID: PMC9671460
- DOI: 10.1371/journal.pcbi.1010591
Probabilistic program inference in network-based epidemiological simulations
Abstract
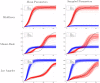
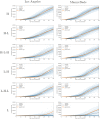
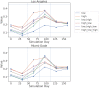
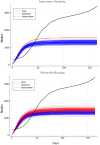
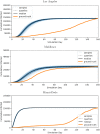
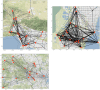
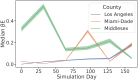
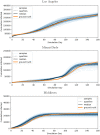
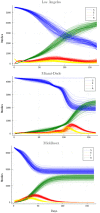
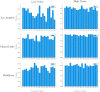
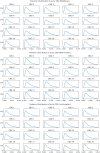
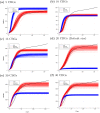
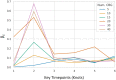
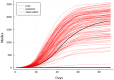
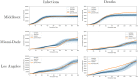
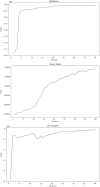
Accurate epidemiological models require parameter estimates that account for mobility patterns and social network structure. We demonstrate the effectiveness of probabilistic programming for parameter inference in these models. We consider an agent-based simulation that represents mobility networks as degree-corrected stochastic block models, whose parameters we estimate from cell phone co-location data. We then use probabilistic program inference methods to approximate the distribution over disease transmission parameters conditioned on reported cases and deaths. Our experiments demonstrate that the resulting models improve the quality of fit in multiple geographies relative to baselines that do not model network topology.
Copyright: © 2022 Smedemark-Margulies et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Tensor product algorithms for inference of contact network from epidemiological data.BMC Bioinformatics. 2024 Sep 2;25(1):285. doi: 10.1186/s12859-024-05910-7. BMC Bioinformatics. 2024. PMID: 39223484 Free PMC article.
-
Stochastic dynamics of an SIS epidemic on networks.J Math Biol. 2022 May 5;84(6):50. doi: 10.1007/s00285-022-01754-y. J Math Biol. 2022. PMID: 35513730
-
Parameter inference in small world network disease models with approximate Bayesian Computational methods.Physica A. 2010 Feb 1;389(3):540-548. doi: 10.1016/j.physa.2009.09.053. Epub 2009 Oct 6. Physica A. 2010. PMID: 32288082 Free PMC article.
-
Probabilistic Models with Deep Neural Networks.Entropy (Basel). 2021 Jan 18;23(1):117. doi: 10.3390/e23010117. Entropy (Basel). 2021. PMID: 33477544 Free PMC article. Review.
-
A tutorial introduction to stochastic simulation algorithms for belief networks.Artif Intell Med. 1993 Aug;5(4):315-40. doi: 10.1016/0933-3657(93)90020-4. Artif Intell Med. 1993. PMID: 8220686 Review.
References
-
- Kermack WO, McKendrick AG. A Contribution to the Mathematical Theory of Epidemics. Proceedings of the Royal Society of London Series A, Containing papers of a mathematical and physical character. 1927;115(772):700–721.
-
- Diekmann O, Heesterbeek JAP. Mathematical Epidemiology of Infectious Diseases: Model Building, Analysis and Interpretation. vol. 5. New York City: John Wiley & Sons; 2000.
-
- Grefenstette JJ, Brown ST, Rosenfeld R, DePasse J, Stone NTB, Cooley PC, et al.. FRED (a Framework for Reconstructing Epidemic Dynamics): An Open-Source Software System for Modeling Infectious Diseases and Control Strategies Using Census-Based Populations. BMC public health. 2013;13:940. doi: 10.1186/1471-2458-13-940 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

