Assessment of genetic markers for multilocus sequence typing (MLST) of Fasciola isolates from Iran
- PMID: 36343016
- PMCID: PMC10029898
- DOI: 10.1002/vms3.995
Assessment of genetic markers for multilocus sequence typing (MLST) of Fasciola isolates from Iran
Abstract
Background: Several markers have been described to characterise the population structure and genetic diversity of Fasciola species (Fasciola hepatica (F. hepatica) and Fasciola gigantica (F. gigantica). However, sequence analysis of a single genomic locus cannot provide sufficient resolution for the genetic diversity of the Fasciola parasite whose genomes are ∼1.3 GB in size.
Objectives: To gain a better understanding of the gene diversity of Fasciola isolates from western Iran and to identify the most informative markers as candidates for epidemiological studies, five housekeeping genes were evaluated using a multilocus sequence typing (MLST) approach.
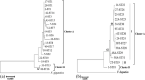
Methods: MLST analysis was developed based on five genes (ND1, Pepck, Pold, Cyt b and HSP70) after genomic DNA extraction, amplification and sequencing. Nucleotide diversity and phylogeny analysis were conducted on both concatenated MLST loci and each individual locus. A median joining haplotype network was created to examine the haplotypes relationship among Fasciola isolates.
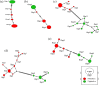
Results: Thirty-three Fasciola isolates (19 F. hepatica and 14 F. gigantica) were included in the study. A total of 2971 bp was analysed for each isolate and 31 sequence types (STs) were identified among the 33 isolates (19 for F. hepatica and 14 for F. gigantica isolates). The STs produced 44 and 42 polymorphic sites and 17 and 14 haplotypes for F. hepatica and F. gigantica, respectively. Haplotype diversity was 0.982 ± 0.026 and 1.000 ± 0.027 and nucleotide diversity was 0.00200 and 0.00353 ± 0.00088 for F. hepatica and F. gigantica, respectively. There was a high degree of genetic diversity with a Simpson's index of diversity of 0.98 and 1 for F. hepatica and F. gigantica, respectively. While HSP70 and Pold haplotypes from Fasciola species were separated by one to three mutational steps, the haplotype networks of ND1 and Cyt b were more complex and numerous mutational steps were found, likely due to recombination.
Conclusions: Although HSP70 and Pold genes from F. gigantica were invariant over the entire region of sequence coverage, MLST was useful for investigating the phylogenetic relationship of Fasciola species. The present study also provided insight into markers more suitable for phylogenetic studies and the genetic structure of Fasciola parasites.
Keywords: Fasciola gigantica; Fasciola hepatica; Iran; genetic diversity; multilocus sequence typing.
© 2022 The Authors. Veterinary Medicine and Science published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures

Similar articles
-
Fasciola spp. in Armenia: Genetic diversity in a global context.Vet Parasitol. 2019 Apr;268:21-31. doi: 10.1016/j.vetpar.2019.02.009. Epub 2019 Mar 1. Vet Parasitol. 2019. PMID: 30981302
-
Morphological and molecular characterization of Fasciola isolates from livestock in Golestan province, northern Iran.Vet Med Sci. 2023 Jul;9(4):1824-1832. doi: 10.1002/vms3.1189. Epub 2023 Jun 15. Vet Med Sci. 2023. PMID: 37317979 Free PMC article.
-
Molecular characterization of Fasciola spp. from the endemic area of northern Iran based on nuclear ribosomal DNA sequences.Exp Parasitol. 2011 Jul;128(3):196-204. doi: 10.1016/j.exppara.2011.03.011. Epub 2011 Apr 2. Exp Parasitol. 2011. PMID: 21440546
-
Chapter 2. Fasciola, lymnaeids and human fascioliasis, with a global overview on disease transmission, epidemiology, evolutionary genetics, molecular epidemiology and control.Adv Parasitol. 2009;69:41-146. doi: 10.1016/S0065-308X(09)69002-3. Adv Parasitol. 2009. PMID: 19622408 Review.
-
Human and Animal Fascioliasis: Origins and Worldwide Evolving Scenario.Clin Microbiol Rev. 2022 Dec 21;35(4):e0008819. doi: 10.1128/cmr.00088-19. Epub 2022 Dec 5. Clin Microbiol Rev. 2022. PMID: 36468877 Free PMC article. Review.
Cited by
-
Real-time impact of COVID-19 pandemic on cutaneous leishmaniasis case finding and strategic planning, preventive interventions, control and epidemiology in a region with a high burden of cutaneous leishmaniasis and COVID-19: A cross-sectional descriptive study based on registry data in Ilam-Iran.Health Sci Rep. 2023 Aug 16;6(8):e1489. doi: 10.1002/hsr2.1489. eCollection 2023 Aug. Health Sci Rep. 2023. PMID: 37599657 Free PMC article.
-
Morphometric and Molecular Characterization of Fasciola spp. in Livestock From Northwestern Provinces of Iran.Vet Med Sci. 2024 Nov;10(6):e70097. doi: 10.1002/vms3.70097. Vet Med Sci. 2024. PMID: 39470121 Free PMC article.
-
Maternal COVID-19 infection and the fetus: Immunological and neurological perspectives.New Microbes New Infect. 2023 Jun;53:101135. doi: 10.1016/j.nmni.2023.101135. Epub 2023 Apr 27. New Microbes New Infect. 2023. PMID: 37143853 Free PMC article. Review.
-
Presence of CRISPR CAS-Like Sequences as a Proposed Mechanism for Horizontal Genetic Exchanges between Trichomonas vaginalis and Its Associated Virus: A Comparative Genomic Analysis with the First Report of a Putative CRISPR CAS Structures in Eukaryotic Cells.Biomed Res Int. 2023 Nov 27;2023:8069559. doi: 10.1155/2023/8069559. eCollection 2023. Biomed Res Int. 2023. PMID: 38058394 Free PMC article.
References
-
- Banu, S. S. , Meyer, W. , Ferreira‐Paim, K. , Wang, Q. , Kuhls, K. , Cupolillo, E. , Schönian, G. , & Lee, R. (2019). A novel multilocus sequence typing scheme identifying genetic diversity amongst Leishmania donovani isolates from a genetically homogeneous population in the Indian subcontinent. International Journal for Parasitology, 49(7), 555–567. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

