Pulmonary Arterial Hypertension Patients Have a Proinflammatory Gut Microbiome and Altered Circulating Microbial Metabolites
- PMID: 36343281
- PMCID: PMC10037487
- DOI: 10.1164/rccm.202203-0490OC
Pulmonary Arterial Hypertension Patients Have a Proinflammatory Gut Microbiome and Altered Circulating Microbial Metabolites
Abstract
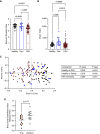
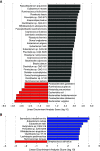
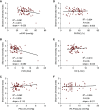
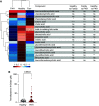
Rationale: Inflammation drives pulmonary arterial hypertension (PAH). Gut dysbiosis causes immune dysregulation and systemic inflammation by altering circulating microbial metabolites; however, little is known about gut dysbiosis and microbial metabolites in PAH. Objectives: To characterize the gut microbiome and microbial metabolites in patients with PAH. Methods: We performed 16S ribosomal RNA gene and shotgun metagenomics sequencing on stool from patients with PAH, family control subjects, and healthy control subjects. We measured markers of inflammation, gut permeability, and microbial metabolites in plasma from patients with PAH, family control subjects, and healthy control subjects. Measurements and Main Results: The gut microbiome was less diverse in patients with PAH. Shannon diversity index correlated with measures of pulmonary vascular disease but not with right ventricular function. Patients with PAH had a distinct gut microbial signature at the phylogenetic level, with fewer copies of gut microbial genes that produce antiinflammatory short-chain fatty acids (SCFAs) and secondary bile acids and lower relative abundances of species encoding these genes. Consistent with the gut microbial changes, patients with PAH had relatively lower plasma concentrations of SCFAs and secondary bile acids. Patients with PAH also had enrichment of species with the microbial genes that encoded the proinflammatory microbial metabolite trimethylamine. The changes in the gut microbiome and circulating microbial metabolites between patients with PAH and family control subjects were not as substantial as the differences between patients with PAH and healthy control subjects. Conclusions: Patients with PAH have proinflammatory gut dysbiosis, in which lower circulating SCFAs and secondary bile acids may facilitate pulmonary vascular disease. These findings support investigating modulation of the gut microbiome as a potential treatment for PAH.
Keywords: dysbiosis; metabolites; microbiome; pulmonary arterial hypertension.
Figures




Comment in
-
Cardiopulmonary Pathogenic Networks: Unveiling the Gut-Lung Microbiome Axis in Pulmonary Arterial Hypertension.Am J Respir Crit Care Med. 2023 Mar 15;207(6):655-657. doi: 10.1164/rccm.202211-2126ED. Am J Respir Crit Care Med. 2023. PMID: 36476165 Free PMC article. No abstract available.
References
-
- Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med . 2016;375:2369–2379. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

