In Multiple Myeloma, High-Risk Secondary Genetic Events Observed at Relapse Are Present From Diagnosis in Tiny, Undetectable Subclonal Populations
- PMID: 36343306
- PMCID: PMC10043564
- DOI: 10.1200/JCO.21.01987
In Multiple Myeloma, High-Risk Secondary Genetic Events Observed at Relapse Are Present From Diagnosis in Tiny, Undetectable Subclonal Populations
Abstract
Purpose: Multiple myeloma (MM) is characterized by copy number abnormalities (CNAs), some of which influence patient outcomes and are sometimes observed only at relapse(s), suggesting their acquisition during tumor evolution. However, the presence of micro-subclones may be missed in bulk analyses. Here, we use single-cell genomics to determine how often these high-risk events are missed at diagnosis and selected at relapse.
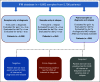
Materials and methods: We analyzed 81 patients with plasma cell dyscrasias using single-cell CNA sequencing. Sixty-six patients were selected at diagnosis, nine at first relapse, and six in presymptomatic stages. A total of 956 newly diagnosed patients with MM and patients with first relapse MM have been identified retrospectively with required cytogenetic data to evaluate enrichment of CNA risk events and survival impact.
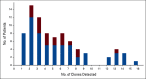
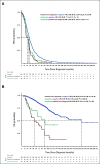
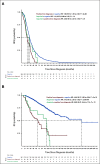
Results: A total of 52,176 MM cells were analyzed. Seventy-four patients (91%) had 2-16 subclones. Among these patients, 28.7% had a subclone with high-risk features (del(17p), del(1p32), and 1q gain) at diagnosis. In a patient with a subclonal 1q gain at diagnosis, we analyzed the diagnosis, postinduction, and first relapse samples, which showed a rise of the high-risk 1q gain subclone (16%, 70%, and 92%, respectively). In our clinical database, we found that the 1q gain frequency increased from 30.2% at diagnosis to 43.6% at relapse (odds ratio, 1.78; 95% CI, 1.58 to 2.00). We subsequently performed survival analyses, which showed that the progression-free and overall survival curves were superimposable between patients who had the 1q gain from diagnosis and those who seemingly acquired it at relapse. This strongly suggests that many patients had 1q gains at diagnosis in microclones that were missed by bulk analyses.
Conclusion: These data suggest that identifying these scarce aggressive cells may necessitate more aggressive treatment as early as diagnosis to prevent them from becoming the dominant clone.
Conflict of interest statement
In Multiple Myeloma, High-Risk Secondary Genetic Events Observed at Relapse are Present From Diagnosis in Tiny, Undetectable Subclonal Populations
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (
Figures


References
-
- Palumbo A, Anderson K: Multiple myeloma. N Engl J Med 364:1046-1060, 2011 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

