Tegument protein UL21 of alpha-herpesvirus inhibits the innate immunity by triggering CGAS degradation through TOLLIP-mediated selective autophagy
- PMID: 36343628
- PMCID: PMC10241001
- DOI: 10.1080/15548627.2022.2139921
Tegument protein UL21 of alpha-herpesvirus inhibits the innate immunity by triggering CGAS degradation through TOLLIP-mediated selective autophagy
Abstract
Alpha-herpesvirus causes lifelong infections and serious diseases in a wide range of hosts and has developed multiple strategies to counteract the host defense. Here, we demonstrate that the tegument protein UL21 (unique long region 21) in pseudorabies virus (PRV) dampens type I interferon signaling by triggering the degradation of CGAS (cyclic GMP-AMP synthase) through the macroautophagy/autophagy-lysosome pathway. Mechanistically, the UL21 protein scaffolds the E3 ligase UBE3C (ubiquitin protein ligase E3C) to catalyze the K27-linked ubiquitination of CGAS at Lys384, which is recognized by the cargo receptor TOLLIP (toll interacting protein) and degraded in the lysosome. Additionally, we show that the N terminus of UL21 in PRV is dominant in destabilizing CGAS-mediated innate immunity. Moreover, viral tegument protein UL21 in herpes simplex virus type 1 (HSV-1) also displays the conserved inhibitory mechanisms. Furthermore, by using PRV, we demonstrate the roles of UL21 in degrading CGAS to promote viral infection in vivo. Altogether, these findings describe a distinct pathway where alpha-herpesvirus exploits TOLLIP-mediated selective autophagy to evade host antiviral immunity, highlighting a new interface of interplay between the host and DNA virus.Abbreviations: 3-MA: 3-methyladenine; ACTB: actin beta; AHV-1: anatid herpesvirus 1; ATG7: autophagy related 7; ATG13: autophagy related 13; ATG101: autophagy related 101; BHV-1: bovine alphaherpesvirus 1; BNIP3L/Nix: BCL2 interacting protein 3 like; CALCOCO2/NDP52: calcium binding and coiled-coil domain 2; CCDC50: coiled-coil domain containing 50; CCT2: chaperonin containing TCP1 subunit 2; CGAS: cyclic GMP-AMP synthase; CHV-2: cercopithecine herpesvirus 2; co-IP: co-immunoprecipitation; CQ: chloroquine; CRISPR: clustered regulatory interspaced short palindromic repeat; Cas9: CRISPR-associated system 9; CTD: C-terminal domain; Ctrl: control; DAPI: 4',6-diamidino-2-phenylindole; DBD: N-terminal DNA binding domain; DMSO: dimethyl sulfoxide; DYNLRB1: dynein light chain roadblock-type 1; EHV-1: equine herpesvirus 1; gB: glycoprotein B; GFP: green fluorescent protein; H&E: hematoxylin and eosin; HSV-1: herpes simplex virus 1; HSV-2: herpes simplex virus 2; IB: immunoblotting; IRF3: interferon regulatory factor 3; lenti: lentivirus; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; MARCHF9: membrane associated ring-CH-type finger 9; MG132: cbz-leu-leu-leucinal; NBR1: NBR1 autophagy cargo receptor; NC: negative control; NEDD4L: NEDD4 like E3 ubiquitin protein ligase; NH4Cl: ammonium chloride; OPTN: optineurin; p-: phosphorylated; PFU: plaque-forming unit; Poly(dA:dT): Poly(deoxyadenylic-deoxythymidylic) acid; PPP1: protein phosphatase 1; PRV: pseudorabies virus; RB1CC1/FIP200: RB1 inducible coiled-coil 1; RNF126: ring finger protein 126; RT-PCR: real-time polymerase chain reaction; sgRNA: single guide RNA; siRNA: small interfering RNA; SQSTM1/p62: sequestosome 1; STING1: stimulator of interferon response cGAMP interactor 1; TBK1: TANK binding kinase 1; TOLLIP: toll interacting protein; TRIM33: tripartite motif containing 33; UL16: unique long region 16; UL21: unique long region 21; UL54: unique long region 54; Ub: ubiquitin; UBE3C: ubiquitin protein ligase E3C; ULK1: unc-51 like autophagy activating kinase 1; Vec: vector; VSV: vesicular stomatitis virus; VZV: varicella-zoster virus; WCL: whole-cell lysate; WT: wild-type; Z-VAD: carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone.
Keywords: Alpha-herpesvirus; CGAS; UL21 protein; selective autophagy; type I interferon signaling.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


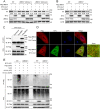

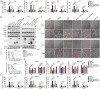
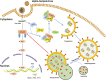
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
