Mitochondrial membrane proteins and VPS35 orchestrate selective removal of mtDNA
- PMID: 36344526
- PMCID: PMC9640553
- DOI: 10.1038/s41467-022-34205-9
Mitochondrial membrane proteins and VPS35 orchestrate selective removal of mtDNA
Abstract
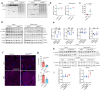
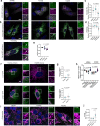
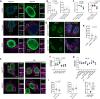
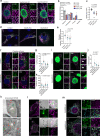
Understanding the mechanisms governing selective turnover of mutation-bearing mtDNA is fundamental to design therapeutic strategies against mtDNA diseases. Here, we show that specific mtDNA damage leads to an exacerbated mtDNA turnover, independent of canonical macroautophagy, but relying on lysosomal function and ATG5. Using proximity labeling and Twinkle as a nucleoid marker, we demonstrate that mtDNA damage induces membrane remodeling and endosomal recruitment in close proximity to mitochondrial nucleoid sub-compartments. Targeting of mitochondrial nucleoids is controlled by the ATAD3-SAMM50 axis, which is disrupted upon mtDNA damage. SAMM50 acts as a gatekeeper, influencing BAK clustering, controlling nucleoid release and facilitating transfer to endosomes. Here, VPS35 mediates maturation of early endosomes to late autophagy vesicles where degradation occurs. In addition, using a mouse model where mtDNA alterations cause impairment of muscle regeneration, we show that stimulation of lysosomal activity by rapamycin, selectively removes mtDNA deletions without affecting mtDNA copy number, ameliorating mitochondrial dysfunction. Taken together, our data demonstrates that upon mtDNA damage, mitochondrial nucleoids are eliminated outside the mitochondrial network through an endosomal-mitophagy pathway. With these results, we unveil the molecular players of a complex mechanism with multiple potential benefits to understand mtDNA related diseases, inherited, acquired or due to normal ageing.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Comment in
-
Endosomal-dependent mitophagy coordinates mitochondrial nucleoid and mtDNA elimination.Autophagy. 2023 Sep;19(9):2609-2610. doi: 10.1080/15548627.2023.2170959. Epub 2023 Jan 29. Autophagy. 2023. PMID: 36691806 Free PMC article.
References
-
- Stewart, J. B. & Chinnery, P. F. Extreme heterogeneity of human mitochondrial DNA from organelles to populations. Nat. Rev. Genet.10.1038/s41576-020-00284-x (2020). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

