Analysis of matched primary and recurrent BRCA1/2 mutation-associated tumors identifies recurrence-specific drivers
- PMID: 36344544
- PMCID: PMC9640723
- DOI: 10.1038/s41467-022-34523-y
Analysis of matched primary and recurrent BRCA1/2 mutation-associated tumors identifies recurrence-specific drivers
Abstract
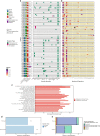
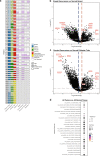
Recurrence is a major cause of death among BRCA1/2 mutation carriers with breast (BrCa) and ovarian cancers (OvCa). Herein we perform multi-omic sequencing on 67 paired primary and recurrent BrCa and OvCa from 27 BRCA1/2 mutation carriers to identify potential recurrence-specific drivers. PARP1 amplifications are identified in recurrences (False Discovery Rate q = 0.05), and PARP1 is significantly overexpressed across primary BrCa and recurrent BrCa and OvCa, independent of amplification status. RNA sequencing analysis finds two BRCA2 isoforms, BRCA2-201/Long and BRCA2-001/Short, respectively predicted to be sensitive and insensitive to nonsense-mediated decay. BRCA2-001/Short is expressed more frequently in recurrences and associated with reduced overall survival in breast cancer (87 vs. 121 months; Hazard Ratio = 2.5 [1.18-5.5]). Loss of heterozygosity (LOH) status is discordant in 25% of patient's primary and recurrent tumors, with switching between both LOH and lack of LOH found. Our study reveals multiple potential drivers of recurrent disease in BRCA1/2 mutation-associated cancer, improving our understanding of tumor evolution and suggesting potential biomarkers.
© 2022. The Author(s).
Conflict of interest statement
S.M.D. has received honoraria from AstraZeneca. J.B. has received honoraria from AstraZeneca and Pfizer. The remaining authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

