Comparative and pangenomic analysis of the genus Streptomyces
- PMID: 36344558
- PMCID: PMC9640686
- DOI: 10.1038/s41598-022-21731-1
Comparative and pangenomic analysis of the genus Streptomyces
Abstract
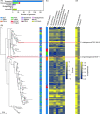
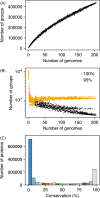
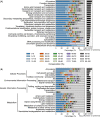
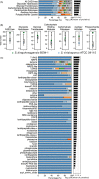
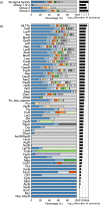
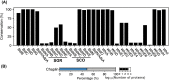
Streptomycetes are highly metabolically gifted bacteria with the abilities to produce bioproducts that have profound economic and societal importance. These bioproducts are produced by metabolic pathways including those for the biosynthesis of secondary metabolites and catabolism of plant biomass constituents. Advancements in genome sequencing technologies have revealed a wealth of untapped metabolic potential from Streptomyces genomes. Here, we report the largest Streptomyces pangenome generated by using 205 complete genomes. Metabolic potentials of the pangenome and individual genomes were analyzed, revealing degrees of conservation of individual metabolic pathways and strains potentially suitable for metabolic engineering. Of them, Streptomyces bingchenggensis was identified as a potent degrader of plant biomass. Polyketide, non-ribosomal peptide, and gamma-butyrolactone biosynthetic enzymes are primarily strain specific while ectoine and some terpene biosynthetic pathways are highly conserved. A large number of transcription factors associated with secondary metabolism are strain-specific while those controlling basic biological processes are highly conserved. Although the majority of genes involved in morphological development are highly conserved, there are strain-specific varieties which may contribute to fine tuning the timing of cellular differentiation. Overall, these results provide insights into the metabolic potential, regulation and physiology of streptomycetes, which will facilitate further exploitation of these important bacteria.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Genome-guided investigation of secondary metabolites produced by a potential new strain Streptomyces BA2 isolated from an endemic plant rhizosphere in Turkey.Arch Microbiol. 2021 Jul;203(5):2431-2438. doi: 10.1007/s00203-021-02210-z. Epub 2021 Mar 5. Arch Microbiol. 2021. PMID: 33666690
-
Insights into Streptomyces spp. isolated from the rhizospheric soil of Panax notoginseng: isolation, antimicrobial activity and biosynthetic potential for polyketides and non-ribosomal peptides.BMC Microbiol. 2020 Jun 3;20(1):143. doi: 10.1186/s12866-020-01832-5. BMC Microbiol. 2020. PMID: 32493249 Free PMC article.
-
Discovery of a new diol-containing polyketide by heterologous expression of a silent biosynthetic gene cluster from Streptomyces lavendulae FRI-5.J Ind Microbiol Biotechnol. 2018 Feb;45(2):77-87. doi: 10.1007/s10295-017-1997-x. Epub 2017 Dec 18. J Ind Microbiol Biotechnol. 2018. PMID: 29255990
-
[Efficient production of polyketide products in Streptomyces hosts - A review].Wei Sheng Wu Xue Bao. 2016 Mar 4;56(3):418-28. Wei Sheng Wu Xue Bao. 2016. PMID: 27382785 Review. Chinese.
-
Genome mining of Streptomyces ambofaciens.J Ind Microbiol Biotechnol. 2014 Feb;41(2):251-63. doi: 10.1007/s10295-013-1379-y. Epub 2013 Nov 21. J Ind Microbiol Biotechnol. 2014. PMID: 24258629 Review.
Cited by
-
PanKB: An interactive microbial pangenome knowledgebase for research, biotechnological innovation, and knowledge mining.Nucleic Acids Res. 2025 Jan 6;53(D1):D806-D818. doi: 10.1093/nar/gkae1042. Nucleic Acids Res. 2025. PMID: 39574409 Free PMC article.
-
Comprehensive analysis of orthologous genes reveals functional dynamics and energy metabolism in the rhizospheric microbiome of Moringa oleifera.Funct Integr Genomics. 2025 Apr 7;25(1):82. doi: 10.1007/s10142-025-01580-7. Funct Integr Genomics. 2025. PMID: 40195156 Free PMC article.
-
Isolation, Genomic Taxonomy, and Genome Characterization of Streptomyces fradiae SS162, Which has Antibacterial Activity Against Vibrio parahaemolyticus.Curr Microbiol. 2025 Aug 6;82(9):441. doi: 10.1007/s00284-025-04426-4. Curr Microbiol. 2025. PMID: 40768039
-
Genomic Diversity of Streptomyces clavuligerus: Implications for Clavulanic Acid Biosynthesis and Industrial Hyperproduction.Int J Mol Sci. 2024 Oct 12;25(20):10992. doi: 10.3390/ijms252010992. Int J Mol Sci. 2024. PMID: 39456781 Free PMC article.
-
Insight into endophytic microbial diversity in two halophytes and plant beneficial attributes of Bacillus swezeyi.Front Microbiol. 2024 Aug 29;15:1447755. doi: 10.3389/fmicb.2024.1447755. eCollection 2024. Front Microbiol. 2024. PMID: 39268535 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

