Machine learning based personalized drug response prediction for lung cancer patients
- PMID: 36344580
- PMCID: PMC9640729
- DOI: 10.1038/s41598-022-23649-0
Machine learning based personalized drug response prediction for lung cancer patients
Abstract
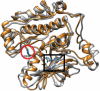
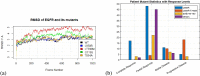
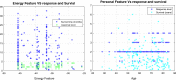
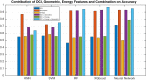
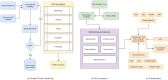
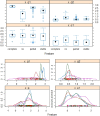
Lung cancers with a mutated epidermal growth factor receptor (EGFR) are a major contributor to cancer fatalities globally. Targeted tyrosine kinase inhibitors (TKIs) have been developed against EGFR and show encouraging results for survival rate and quality of life. However, drug resistance may affect treatment plans and treatment efficacy may be lost after about a year. Predicting the response to EGFR-TKIs for EGFR-mutated lung cancer patients is a key research area. In this study, we propose a personalized drug response prediction model (PDRP), based on molecular dynamics simulations and machine learning, to predict the response of first generation FDA-approved small molecule EGFR-TKIs, Gefitinib/Erlotinib, in lung cancer patients. The patient's mutation status is taken into consideration in molecular dynamics (MD) simulation. Each patient's unique mutation status was modeled considering MD simulation to extract molecular-level geometric features. Moreover, additional clinical features were incorporated into machine learning model for drug response prediction. The complete feature set includes demographic and clinical information (DCI), geometrical properties of the drug-target binding site, and the binding free energy of the drug-target complex from the MD simulation. PDRP incorporates an XGBoost classifier, which achieves state-of-the-art performance with 97.5% accuracy, 93% recall, 96.5% precision, and 94% F1-score, for a 4-class drug response prediction task. We found that modeling the geometry of the binding pocket combined with binding free energy is a good predictor for drug response. However, we observed that clinical information had a little impact on the performance of the model. The proposed model could be tested on other types of cancers. We believe PDRP will support the planning of effective treatment regimes based on clinical-genomic information. The source code and related files are available on GitHub at: https://github.com/rizwanqureshi123/PDRP/ .
© 2022. The Author(s).
Figures

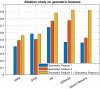
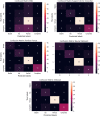
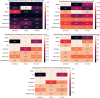
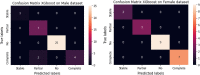

Similar articles
-
Computational Methods for the Analysis and Prediction of EGFR-Mutated Lung Cancer Drug Resistance: Recent Advances in Drug Design, Challenges and Future Prospects.IEEE/ACM Trans Comput Biol Bioinform. 2023 Jan-Feb;20(1):238-255. doi: 10.1109/TCBB.2022.3141697. Epub 2023 Feb 3. IEEE/ACM Trans Comput Biol Bioinform. 2023. PMID: 35007197 Review.
-
Contribution of EGFR and ErbB-3 Heterodimerization to the EGFR Mutation-Induced Gefitinib- and Erlotinib-Resistance in Non-Small-Cell Lung Carcinoma Treatments.PLoS One. 2015 May 20;10(5):e0128360. doi: 10.1371/journal.pone.0128360. eCollection 2015. PLoS One. 2015. PMID: 25993617 Free PMC article.
-
Epidermal growth factor receptor (EGFR) mutations in a series of non-small-cell lung cancer (NSCLC) patients and response rate to EGFR-specific tyrosine kinase inhibitors (TKIs).Clin Transl Oncol. 2011 Nov;13(11):812-8. doi: 10.1007/s12094-011-0739-1. Clin Transl Oncol. 2011. PMID: 22082647
-
Optimizing the sequencing of tyrosine kinase inhibitors (TKIs) in epidermal growth factor receptor (EGFR) mutation-positive non-small cell lung cancer (NSCLC).Lung Cancer. 2019 Nov;137:113-122. doi: 10.1016/j.lungcan.2019.09.017. Epub 2019 Sep 23. Lung Cancer. 2019. PMID: 31568888 Free PMC article. Review.
-
Lung Cancer Driven by BRAFG469V Mutation Is Targetable by EGFR Kinase Inhibitors.J Thorac Oncol. 2022 Feb;17(2):277-288. doi: 10.1016/j.jtho.2021.09.008. Epub 2021 Oct 12. J Thorac Oncol. 2022. PMID: 34648945
Cited by
-
Research progress of HIF-1a on immunotherapy outcomes in immune vascular microenvironment.Front Immunol. 2025 Feb 6;16:1549276. doi: 10.3389/fimmu.2025.1549276. eCollection 2025. Front Immunol. 2025. PMID: 39981236 Free PMC article. Review.
-
Artificial intelligence and anti-cancer drugs' response.Acta Pharm Sin B. 2025 Jul;15(7):3355-3371. doi: 10.1016/j.apsb.2025.05.009. Epub 2025 May 21. Acta Pharm Sin B. 2025. PMID: 40698144 Free PMC article. Review.
-
Predicting the hub interactome of COVID-19 and oral squamous cell carcinoma: uncovering ALDH-mediated Wnt/β-catenin pathway activation via salivary inflammatory proteins.Sci Rep. 2025 Feb 3;15(1):4068. doi: 10.1038/s41598-025-88819-2. Sci Rep. 2025. PMID: 39901050 Free PMC article.
-
Integrative machine learning approach for forecasting lung cancer chemosensitivity: From algorithm to cell line validation.Comput Struct Biotechnol J. 2025 Jul 24;27:3307-3318. doi: 10.1016/j.csbj.2025.07.043. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40778317 Free PMC article.
-
Preclinical Anticipation of On- and Off-Target Resistance Mechanisms to Anti-Cancer Drugs: A Systematic Review.Int J Mol Sci. 2024 Jan 5;25(2):705. doi: 10.3390/ijms25020705. Int J Mol Sci. 2024. PMID: 38255778 Free PMC article.
References
-
- Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. (2022). - PubMed
-
- Gupta GP, Massagué J. Cancer metastasis: Building a framework. Cell. 2006;127:679–695. - PubMed
-
- Qureshi, R. et al. Computational methods for the analysis and prediction of egfr-mutated lung cancer drug resistance: Recent advances in drug design, challenges and future prospects. IEEE/ACM Trans. Comput. Biol. Bioinform. (2022). - PubMed
-
- Kawaguchi T, et al. Randomized phase iii trial of erlotinib versus docetaxel as second-or third-line therapy in patients with advanced non-small-cell lung cancer: Docetaxel and erlotinib lung cancer trial (delta) J. Clin. Oncol. 2014;32:1902–1908. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

