Probabilistic model checking of cancer metabolism
- PMID: 36344581
- PMCID: PMC9640632
- DOI: 10.1038/s41598-022-21846-5
Probabilistic model checking of cancer metabolism
Abstract
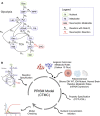
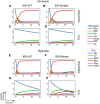
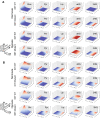
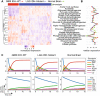
Cancer cell metabolism is often deregulated as a result of adaption to meeting energy and biosynthesis demands of rapid growth or direct mutation of key metabolic enzymes. Better understanding of such deregulation can provide new insights on targetable vulnerabilities, but is complicated by the difficulty in probing cell metabolism at different levels of resolution and under different experimental conditions. We construct computational models of glucose and glutamine metabolism with focus on the effect of IDH1/2-mutations in cancer using a combination of experimental metabolic flux data and patient-derived gene expression data. Our models demonstrate the potential of computational exploration to reveal biologic behavior: they show that an exogenously-mutated IDH1 experimental model utilizes glutamine as an alternative carbon source for lactate production under hypoxia, but does not fully-recapitulate the patient phenotype under normoxia. We also demonstrate the utility of using gene expression data as a proxy for relative differences in metabolic activity. We use the approach of probabilistic model checking and the freely-available Probabilistic Symbolic Model Checker to construct and reason about model behavior.
© 2022. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

